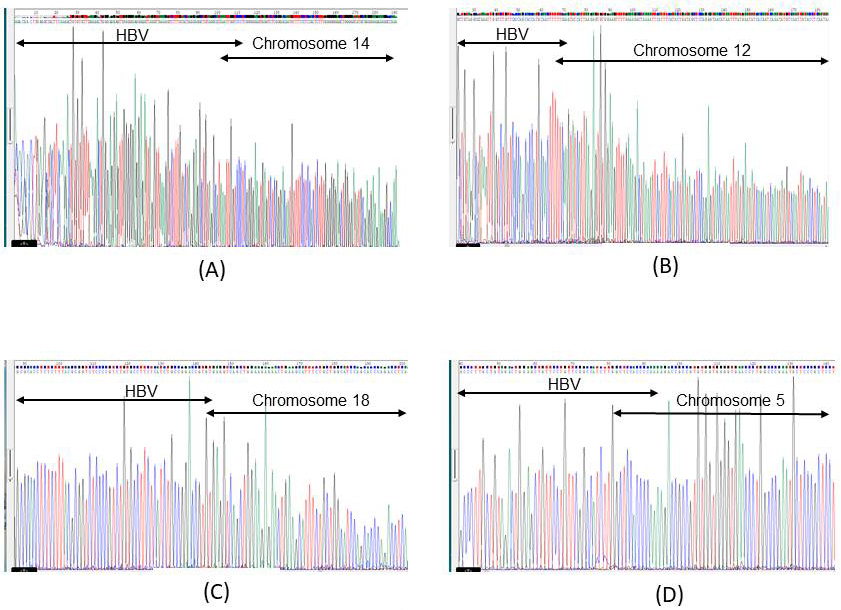
Figure 1. Electropherogram of chimeric reads by Sanger sequencing
A) Ig18206 liver. Electropherogram of the conventional PCR results for chromosome 14: 68,792,458 +HBV (total read number: 7 reads). The chimeric read detected by capture-based next-generation sequencing (NGS) is as follows:
TTGTACTAGGAGGCTGTAGGCATAAATTGGTCCCTCGGGGAAGTGAGTCTCGGGACAATGTTTTCCTCCACTCTTTGGGGGGAGCTGGGGATATGCGA
B) Ig18206 liver. Electropherogram of the conventional PCR results for chromosome 12: 95,875,424 +HBV (total read number: 1 read). The chimeric read detected by capture-based NGS is as follows:
GGCATAAATTGGTCTGTTCACCAGCACCATGCAACTTTTTCAAGTATCATTAAGAGTGTGGAAGTTTTGAAAGACTAAAATTCATTTCATACTGATAGCTC
C) Ig18807 liver. Electropherogram of the conventional PCR results for chromosome 18: 59,672,521 +HBV (total read number: 73 reads). The chimeric read detected by capture-based NGS is as follows:
CACGGGGCGCACCTCTCTTTACGCGGTCTCCCCGTCTGTGCCTTCTCATCTGCCGGACCGTGTGAGGTCAGCTGGAAGAGAACTGAAGCATTTCTGCTGGCATCCAGCACCCAGACCTCAGACACAAGAGTGAGGCCCT
D) Ig18207 whole blood cells. Electropherogram of the conventional PCR results for chromosome 5: 44,013,798, +HBV (total read number: 1 read). The chimeric read detected by capture-based NGS is as follows:
GGAGCATTCGGGCCAGGGTTCACCCCACCACACGGCGGTCTTTTGGGGTGGAGCCAAGATGGACAAATAGGAACAGCTCCAGTCTACAGCTCCCAGCTTGA
However, Sanger sequencing showed that the integration breakpoint of the chimeric read was chromosome 5: 75,175,747. The chimeric read detected by Sanger sequencing is as follows:
TCAAATCAGGTAGGAGCGGGAGCATTCGGGCCAGGGTTCACCCCACCACACGGCGGTCTTTTGGGGTGGAGCCAAGATGGCCGAACAGGAACAGCTCCAGTCTACAGC
The underline indicates the difference in nucleotides between next-generation and Sanger sequencing.
From: Integration of Viral Genome to Human Genomic DNA in Nails of Patients with Chronic Hepatitis B Virus Infection