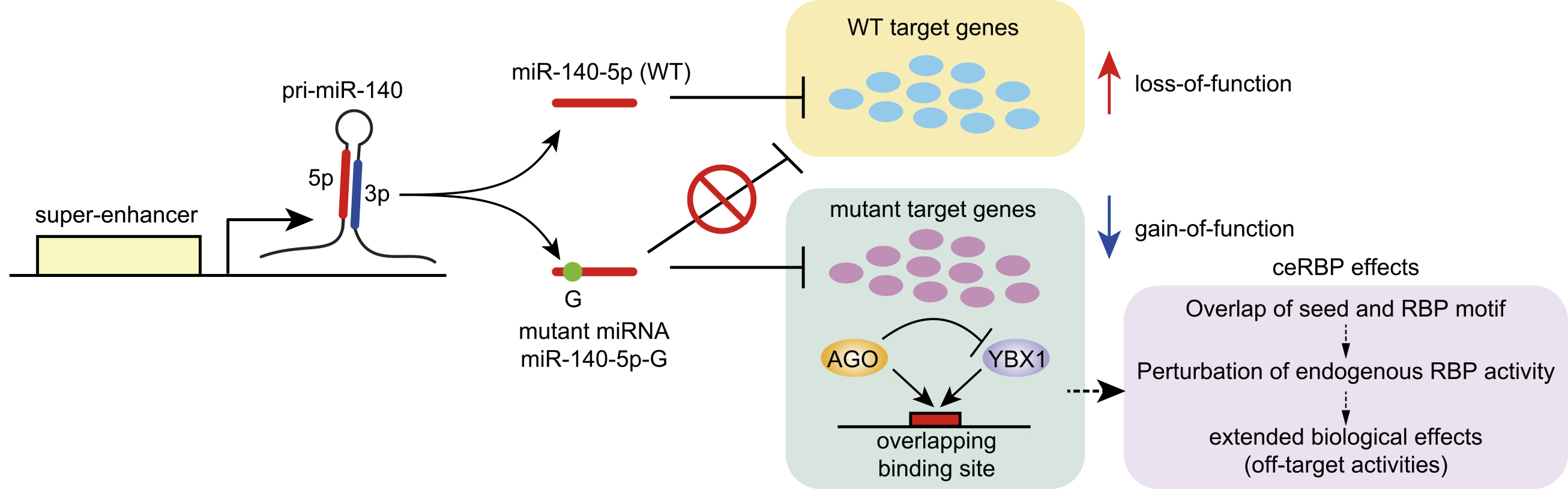
Figure 1. Overview of the miRNA biogenesis pathway in mammalian cells.
The canonical miRNA biogenesis pathway includes multiple steps, including the transcription of pri-miRNAs, DROSHA-mediated processing in the nucleus, export to the cytoplasm, DICER-mediated processing in the cytoplasm, loading onto AGO proteins, and RISC formation. Several sources of noncanonical miRNAs that are generated independently of DROSHA or DICER are known. Erythrocyte-specific miR-451 is generated via the cluster assistance mechanism of DROSHA processing and DICER-independent AGO2-mediated processing.
From: Roles of MicroRNAs in Disease Biology
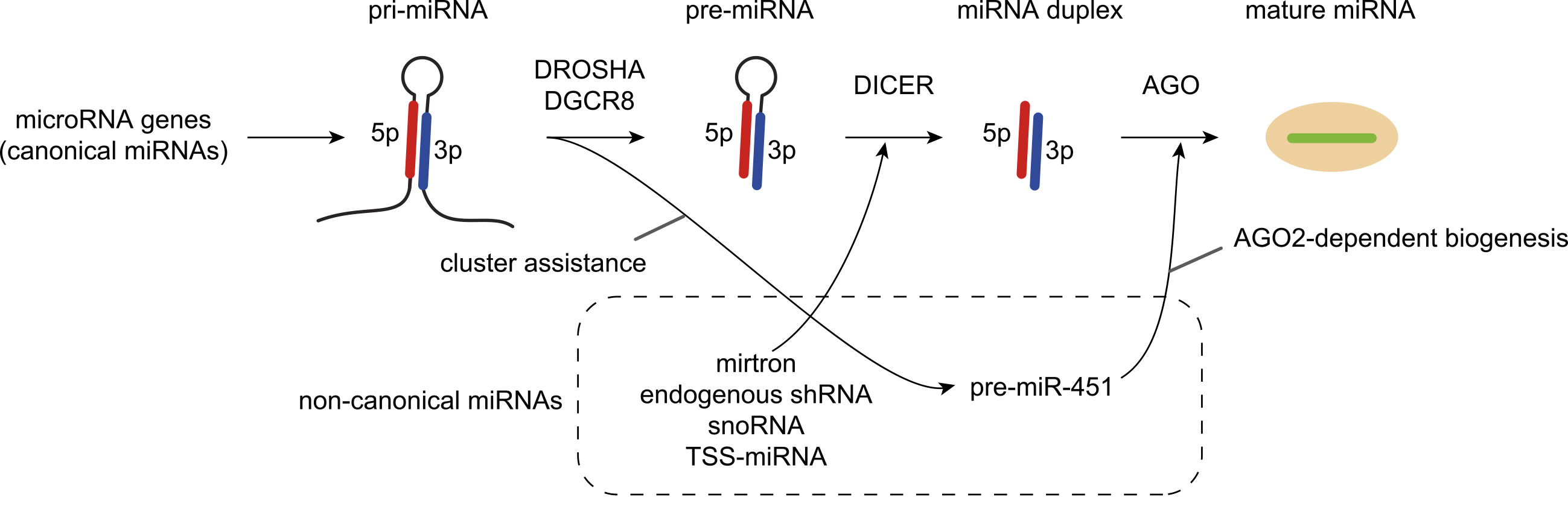
Figure 2. Effects of miRNA-mRNA interactions on target regulation and miRNA stability.
a, Classification of canonical miRNA target sites.
b, Effects of multiple miRNA target sites and extensive miRNA-mRNA target interactions on target repression and miRNA stability.
From: Roles of MicroRNAs in Disease Biology
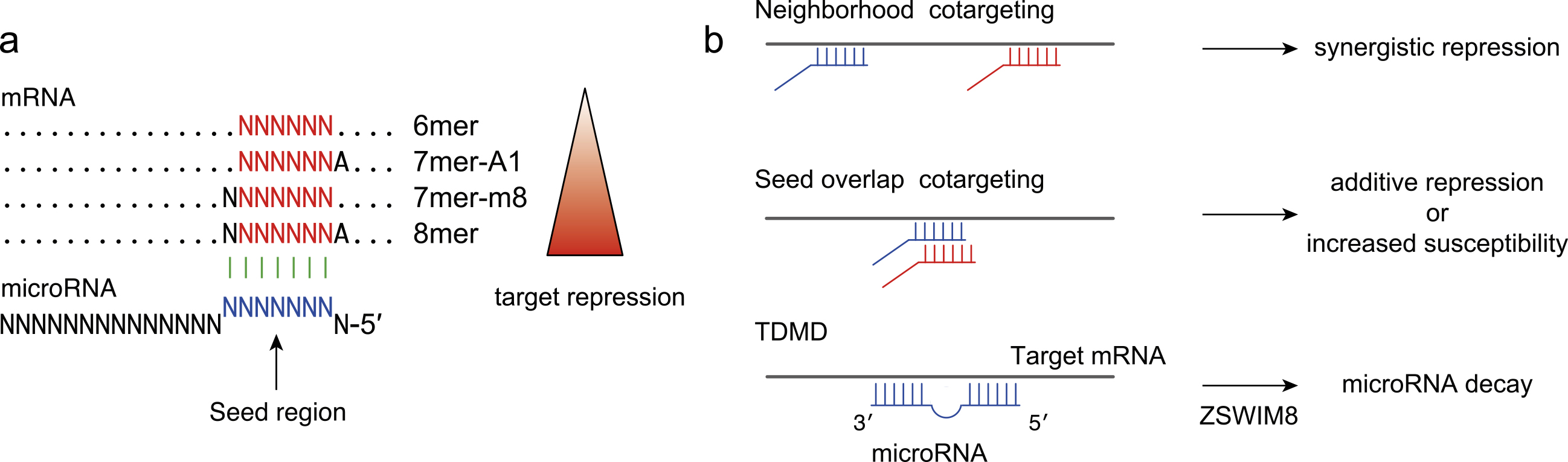
Figure 3. Mechanisms of miRNA dysregulation in cancer.
a, The altered expression of tumor-suppressive or oncogenic miRNAs is induced by various mechanisms: (1) chromosomal abnormalities; (2) alterations in transcriptional regulation; (3) epigenetic mechanisms including super-enhancers; (4) mutations or modifications in miRNAs; and (5) dysregulation of the miRNA biogenesis pathway.
b, Dysregulation of DICER in DICER1 syndrome. Several models of two-hit activation are shown. Various other scenarios have also been reported.
c, Mutations in DROSHA and DGCR8 in Wilms tumors.
From: Roles of MicroRNAs in Disease Biology
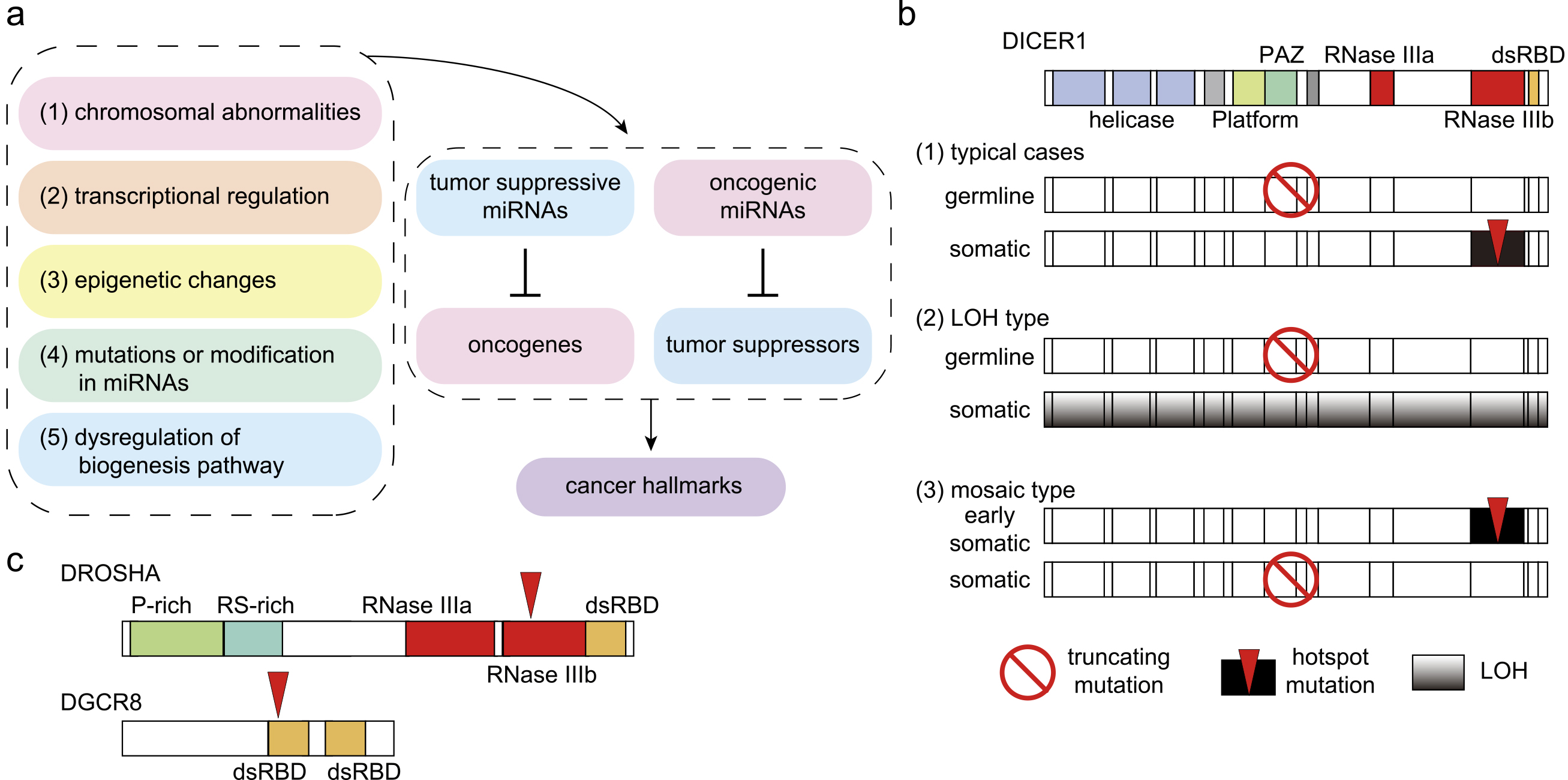
Figure 4. Pathogenic mutation of miR-140 in skeletal dysplasia and ceRBP effects.
(Cited and modified from Grigelioniene G, Suzuki HI, Taylan F, et al. Gain-of-function mutation of microRNA-140 in human skeletal dysplasia. Nat Med. 2019;25(4):583-90(75) and Suzuki HI, Spengler RM, Grigelioniene G, et al. Deconvolution of seed and RNA-binding protein crosstalk in RNAi-based functional genomics. Nat Genet. 2018;50(5):657-61(76). Copyright of the figure belongs to the authors.)
A seed region mutation in chondrocyte-specific miR-140 has been identified in spondyloepiphyseal dysplasia MIR140 type Nishimura (OMIM #618618). This mutation results in widespread derepression of wild-type miR-140-5p targets and repression of mutant miR-140-5p (miR-140-5p-G) targets. Additionally, the miR-140-5p mutant seed competes with the Ybx1 RNA-binding protein for the overlapping binding sites.
From: Roles of MicroRNAs in Disease Biology