Corresponding author: Hiroshi Yokomichi, hyokomichi@yamanashi.ac.jp
DOI: 10.31662/jmaj.2020-0016
Received: April 7, 2020
Accepted: May 1, 2020
Advance Publication: July 7, 2020
Published: July 15, 2020
Cite this article as:
Yokomichi H, Inozume T, Wada M, Asai J, Igaki H, Namikawa K, Hayashi A, Fukushima S, Fujimura T, Koga H, Nakamura Y, Mochizuki M, Yamagata Z. Concordance and Discordance Rates of V-Raf Murine Sarcoma Viral Oncogene Homolog B1 (BRAF)V600E Status in Metastatic against Primary Lesion of Melanoma: A Meta-analysis. JMA J. 2020;3(3):274-279.
Key words: melanoma, BRAFV600E, primary tumor, metastasis, BRAF mutation, disagreement, serine-threonine protein kinase BRAF inhibitor, mitogen-activated protein kinase kinase inhibitor
New effective molecular-targeted agents (1) including combination serine-threonine protein kinase BRAF inhibitor and mitogen-activated protein kinase kinase (MEK) inhibitor (BRAF/MEK inhibitor) therapy have improved overall survival (OS) and progression-free survival (PFS) of patients with metastatic melanoma with BRAFV600E/K mutations (2), (3), (4). Observational studies suggest that 40%-60% Caucasians and < 30% Japanese are BRAFV600E/K mutation positive (5). BRAF/MEK inhibitors are only approved for BRAFV600E/K mutation-positive patients because they are only effective against lesions with this mutation (6).
The mutational status of metastatic lesions cannot easily be tested because melanoma metastasizes to subcutaneous and superficial lymph nodes, visceral areas, and deep lymph nodes; therefore, in such cases, whether BRAF/MEK inhibitors are suitable for patients with melanoma is unclear (7). In clinical practice, primary skin lesions resected during primary treatment are genetically tested to predict the mutational status of inaccessible metastatic lesions.
Some cases might have BRAFV600E/K mutation-positive primary lesions, but have negative metastatic lesions, or vice versa. In the former, BRAF/MEK inhibitors would not be effective, whereas in the latter, the patient loses the opportunity to receive effective treatment. Disagreement in the proportion of BRAF mutations between primary and metastatic lesions varies (8). A previous study did not address the probability of agreement for metastasis when the primary lesion was BRAF mutation positive or BRAF mutation negative. Moreover, disagreement proportions for all BRAF mutations were included, although molecular-targeted therapy is currently approved only for patients with BRAFV600E/K (8).
Calculating the mutation probability in metastatic lesions when the primary lesion mutation is known allows the more appropriate use of BRAF/MEK inhibitors. The probability of BRAFV600E-positive [BRAF(+)] metastatic lesions when the primary cancer lesion was BRAF(+) and BRAFV600E-negative [BRAF(−)] metastatic lesions when the primary cancer lesion was BRAF(−) was calculated.
We searched the PubMed, Cochrane Library, and Igaku Chuo Zasshi databases for observational studies on November 23, 2017. Search criteria with MeSH terms were published elsewhere (9). One investigator selected articles that potentially met the criteria on the basis of their titles and abstracts. For eligible studies, the same investigator abstracted the data independently using a predefined form. Searched studies used real-time PCR, Sanger sequencing, and BRAFV600E-specific immunohistochemistry to detect mutants. Therefore, we set the outcome of BRAFV600E mutations as those detected by the searched methods and that were predominant in 79%-90% of patients (10), (11). We included studies investigating BRAFV600E status between primary melanoma and metastatic lesions in the same patients, irrespective of testing methods, and those without BRAFV600E testing were excluded.
We integrated the probabilities of BRAFV600E positive and negative in metastatic lesions when primary lesions were BRAFV600E positive and negative, respectively: BRAF(+) metastatic lesion given the primary lesion is BRAF(+), and BRAF(−) metastatic lesion given the primary lesion is BRAF(−). To determine these probabilities, we used primary/metastatic lesion [mutation positive/negative, represented as BRAF(+) or (−)/BRAF(+) or (−)] information for each individual. We calculated the integrated probability (95% confidence interval [95%CI]) of BRAF(+) metastatic lesions given BRAF(+) primary lesions and that of BRAF(−) metastatic lesions given BRAF(−) primary lesions. To unite disagreements in proportions between test results of primary and metastatic lesions, we included more studies. In this analysis, detailed test results for primary and metastatic lesions were not required, but cross tabulations of BRAF(+)/(+), BRAF(+)/(−), BRAF(−)/(+), and BRAF(−)/(−) from total sample populations in each observational study were needed. We integrated the proportions of disagreement for the numbers of BRAF(+)/(−) or BRAF(−)/(+) cases divided by the numbers of BRAF(+)/(+), BRAF(+)/(−), BRAF(−)/(+), and BRAF(−)/(−) cases (all combinations of positive/negative results in primary and metastatic lesions). We presented the proportion of disagreement for the number of discrepant cases divided by the number of consistent plus discrepant cases.
To integrate the probabilities and proportions, we used DerSimonian-Laird estimation (random effect model) and fixed-effects models for meta-analysis. Results are shown as forest plots, and publication bias was checked by funnel plots. We used R x64 V.4.4.2 software for statistical analyses and illustrations. All P-values are two-sided, and P < 0.05 indicated statistical significance.
This study was approved by the ethics committee of the School of Medicine, University of Yamanashi (approval number: 1894), in accordance with the ethical guidelines and regulations of the Declaration of Helsinki.
Supplementary Figure 1 shows our literature searching and screening processes. Thirteen studies were included in the integration of probabilities of agreement (12), (13), (14), (15), (16), (17), (18), (19), (20), (21), (22), (23), (24). Figure 1 shows the probabilities of BRAF(+) metastatic lesions given BRAF(+) primary lesions: the integrated probability is 0.84 (95%CI: 0.79-0.90) in the fixed-effects model and 0.82 (95%CI: 0.71-0.94) in the random-effects model. Figure 2 shows the probabilities of BRAF(−) metastatic lesions with BRAF(−) primary lesions: the integrated probability is 0.86 (95%CI: 0.80-0.91) in the fixed-effects model and 0.82 (95%CI: 0.70-0.94) in the random-effects model. For the integration of disagreement proportions between primary and metastatic lesions with BRAFV600E, 20 studies (12), (13), (14), (15), (16), (17), (18), (19), (20), (21), (22), (23), (24), (25), (26), (27), (28), (29), (30), (31) were included. Figure 3 shows the integrated proportions of 0.13 (95%CI: 0.11-0.16) in the fixed-effects model and 0.13 (95%CI: 0.08-0.18) in the random-effects model. I2 indexes for heterogeneity were 76%, 80%, and 72% in Figure 1, Figure 2, and Figure 3, respectively. Funnel plots (Supplementary Figure 2, Figure 3, and Figure 4) indicated no publication bias.
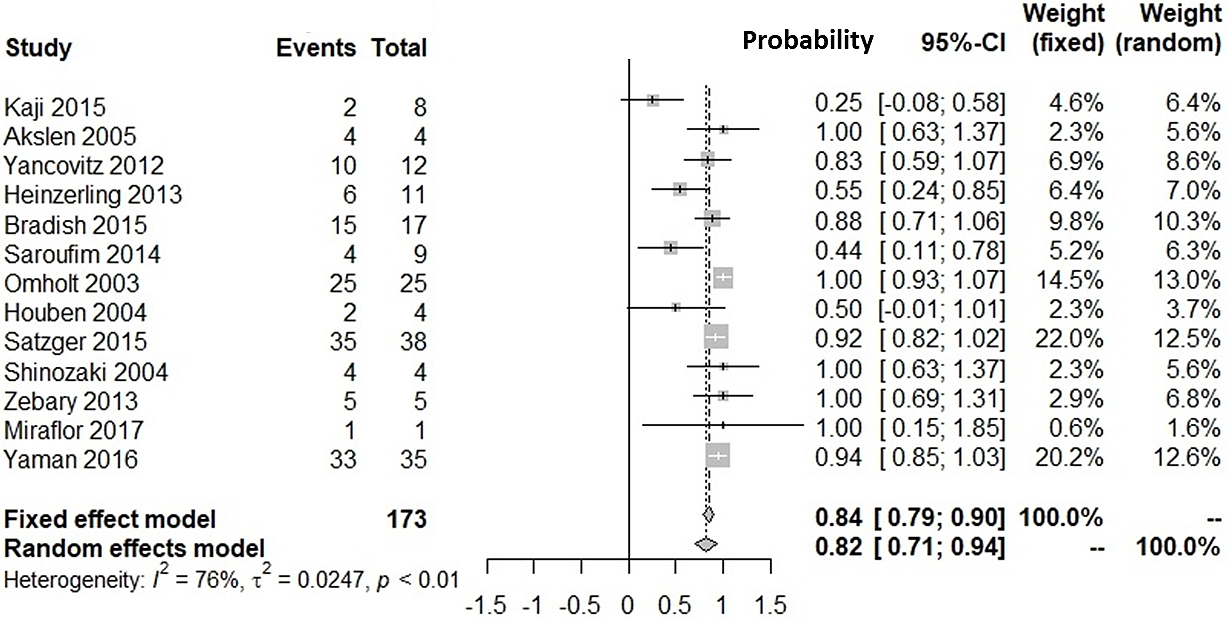
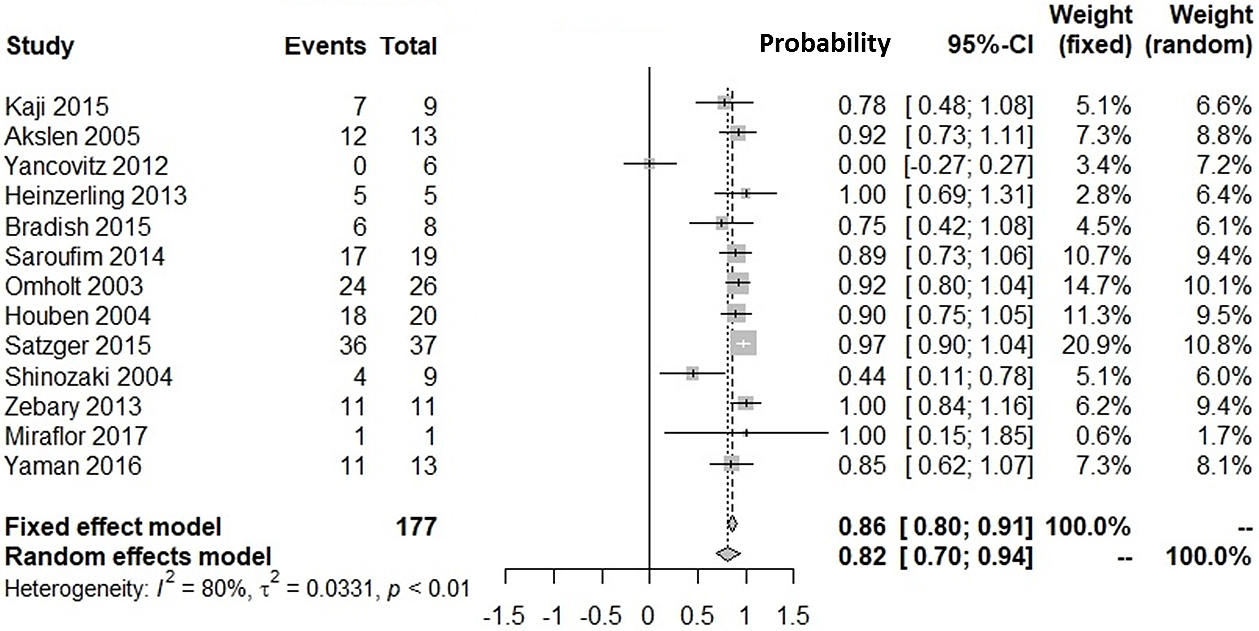
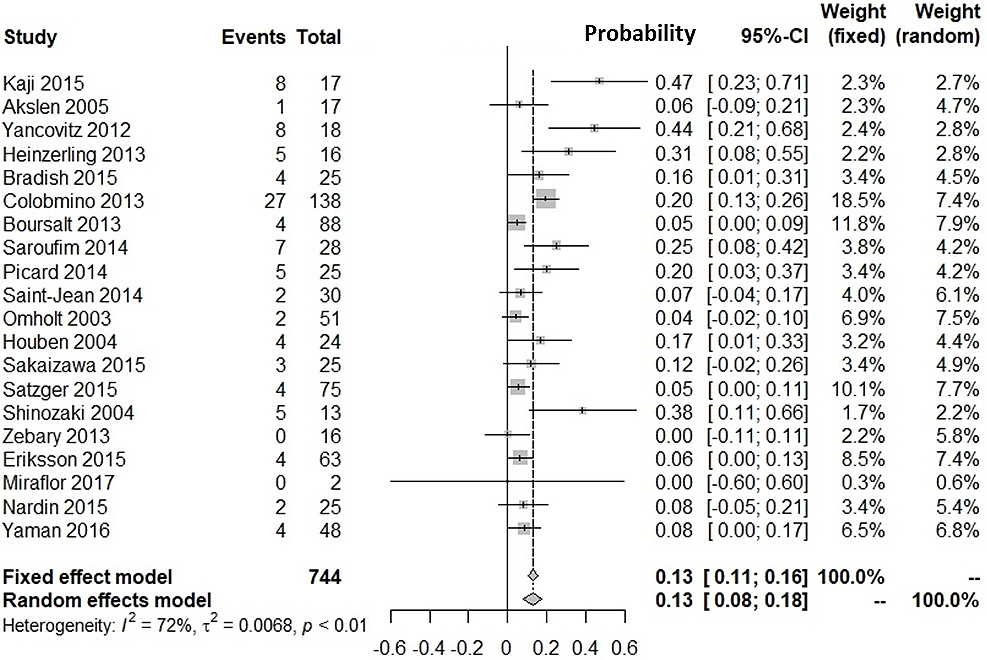
We investigated the probability of BRAFV600E-positive metastases against BRAFV600E-positive primary lesions and that of BRAFV600E-negative metastases against BRAFV600E-negative primary lesions. Our meta-analyses showed the probability of BRAF(+) metastatic lesions with BRAF(+) primary lesions was 82% (95%CI: 71%-94%). The probability of BRAF(−) metastatic lesions with BRAF(−) primary lesions was 82% (95%CI: 70%-94%). The proportion of discrepancies between primary and metastatic lesions was 13% (95%CI: 8%-18%). The proportion was similar with that for epidermal growth factor receptor mutation in patients with non-small cell lung cancer (12.2%) (32).
BRAF/MEK inhibitors have changed metastatic melanoma treatment. In the beginning, BRAF inhibitor monotherapy has improved OS and PFS in BRAFV600E/K patients (33), (34). Thereafter, randomized controlled trials showed combination therapy with BRAF/MEK inhibitors was superior to BRAF inhibitor monotherapy regarding OS and PFS (2). Moreover, a lower frequency of adverse events (rash, alopecia, and skin tumors) was observed for combination therapy compared with that for monotherapy (2), (3). Thus, combination therapy has become a standard approach (7).
BRAFV600E/K testing in primary lesions is critical for patients with metastasis who may respond to BRAF/MEK inhibitors (35). Currently, agents are usually administered to patients according to genetic tests performed on primary lesion tissues. Our findings might convince clinicians that metastatic lesions in 82%-87% of patients with BRAFV600E-positive primary lesions are sensitive to BRAF/MEK inhibitors (Figure 1, Figure 3), whereas 13%-18% would not show an initial response to therapy in clinical trials. Therefore, clinicians might be encouraged to test for BRAF mutations in metastatic lesions, because patients with discrepant results between primary and metastatic lesions (14%-18% probability) are often disadvantaged in relation to treatment decisions.
Our study had several strengths. We accumulated the results of 20 worldwide studies, including 15 in a previous meta-analysis of disagreement (8). We focused on BRAFV600E, the most frequent and clinically important mutation in melanoma. Funnel plots indicated collated data had no publication bias.
The present study also had some limitations. Selection bias was possible; collated data were from patients who underwent genetic tests, which might decrease the generalizability of the results. Genetic testing methods varied among studies. Ethnic background and stage of melanoma varied and should be considered for treatment decisions after genetic testing. High heterogeneity (I2 = 72%-80%) was observed in this meta-analysis. Therefore, the agreement and disagreement proportions might vary among regions.
In conclusion, this meta-analysis revealed 82% of patients had BRAFV600E-positive metastatic lesions with BRAFV600E-positive primary lesions and 82% of patients with BRAFV600E-negative primary lesions had BRAFV600E-negative metastatic lesions. The proportion of disagreement between BRAFV600E mutations in primary and metastatic lesions was 13%. Over-reliance on genetic results of primary tumors might prevent patients with discrepant results receiving appropriate treatment for metastatic lesions. Genetic testing for BRAFV600 mutations using metastatic tumor samples is suggested, if available, without invasive biopsies.
KN received honoraria from Ono Pharmaceutical, Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis Pharmaceutical, Toray Industries, and Takara Bio, outside the submitted work; YN received honoraria from Ono Pharmaceutical, Bristol-Myers Squibb, Maruho, Kyowa Kirin, Merck Sharp & Dohme, Novartis Pharmaceutical, Taisho Toyama Pharma, and Taiho Pharma, outside the submitted work.
This work was partly supported by the Japan Society for the Promotion of Science grant (KAKENHI Grant Number JP18K17376) to HY. The funding source had no role in the design of this study and will not have any role during its execution, analyses, interpretation of the data, or decision to submit results.
We thank Nikki March, PhD, for editing a draft of this manuscript.
Conception/design: Inozume T, Yokomichi H
Acquisition of data: All authors
Data analysis: Yokomichi H
Interpretation: All authors
Manuscript writing: Inozume T, Yokomichi H
Final approval of manuscript: All authors
Hiroshi Yokomichi and Takashi Inozume contributed equally to this work.
This study was approved by the ethics committee of the School of Medicine, University of Yamanashi (approval number: 1894), in accordance with the ethical guidelines and regulations of the Declaration of Helsinki.
Zentaro Yamagata is one of the Editors of JMA Journal and on the journal's Editorial Staff. He was not involved in the editorial evaluation or decision to accept this article for publication at all.
Li F, Zhao C, Wang L. Molecular-targeted agents combination therapy for cancer: developments and potentials. Int J Cancer. 2014;134(6):1257-69.
Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694-703.
Robert C, Karaszewska B, Schachter J, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30-9.
Robert C, Grob JJ, Stroyakovskiy D, et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med. 2019;381(7):626-36.
Ashida A, Uhara H, Kiniwa Y, et al. Assessment of BRAF and KIT mutations in Japanese melanoma patients. J Dermatol Sci. 2012;66(3):240-2.
Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105(8):3041-6.
Nakamura Y, Asai J, Igaki H, et al. Japanese Dermatological Association Guidelines: Outlines of guidelines for cutaneous melanoma 2019. J Dermatol. 2020;47(2):89-103.
Vachis A, Ullenhag GJ. Discrepancy in BRAF status among patients with metastatic malignant melanoma: A meta-analysis. Eur J Cancer. 2017;81:106-15.
Nakamura Y, Asai J, Igaki H, et al. Japanese Dermatological Association Guidelines, 3rd ed: Guidelines for cutaneous melanoma 2019. Nihon Hifuka Gakkai Zasshi. 2019;9(129):1759-843. Japanese.
Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507-16.
Lovly CM, Dahlman KB, Fohn LE, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic rials. PLoS One. 2012;7(4):e35309.
Kaji T, Yamasaki O, Takata M, et al. Comparative study on driver mutations in primary and metastatic melanomas at a single Japanese institute: a clue for intra-and inter-tumor heterogeneity. J Dermatol Sci. 2017;85(1):51-7.
Akslen LA, Angelini S, Straume O, et al. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J Gen Intern Med. 2005;20(5):312-7.
Yancovitz M, Litterman A, Yoon J, et al. Intra-and inter-tumor heterogeneity of BRAFV600Emutations in primary and metastatic melanoma. PLoS One. 2012;7(1):e29336.
Heinzerling L, Baiter M, Kühnapfel S, et al. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br J Cancer. 2013;109(11):2833-41.
Bradish JR, Richey JD, Post KM, et al. Discordancy in BRAF mutations among primary and metastatic melanoma lesions: clinical implications for targeted therapy. Mod Pathol. 2015;28(4):480-6.
Saroufim M, Habib RH, Gerges R, et al. Comparing BRAF mutation status in matched primary and metastatic cutaneous melanomas: implications on optimized targeted therapy. Exp Mol Pathol. 2014;97(3):315-20.
Omholt K, Platz A, Kanter L, et al. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9(17):6483-8.
Houben R, Becker JC, Kappel A, et al. Constitutive activation of the Ras-Raf signaling pathway in metastatic melanoma is associated with poor prognosis. J Carcinog. 2004;3:6.
Satzger I, Marks L, Kerick M, et al. Allele frequencies of BRAFV600 mutations in primary melanomas and matched metastases and their relevance for BRAF inhibitor therapy in metastatic melanoma. Oncotarget. 2015;6(35):37895.
Shinozaki M, Fujimoto A, Morton DL, et al. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10(5):1753-7.
Zebary A, Omholt K, Vassilaki I, et al. KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J Dermatol Sci. 2013;72(3):284-9.
Miraflor AP, de Abreu FB, Peterson JD, et al. Somatic mutation analysis in melanoma using targeted next generation sequencing. Exp Mol Pathol. 2017;103(2):172-7.
Yaman B, Kandiloglu G, Akalin T. BRAF-V600 mutation heterogeneity in primary and metastatic melanoma: a study with pyrosequencing and immunohistochemistry. Am J Dermatopathol. 2016;38(2):113-20.
Colombino M, Lissia A, Capone M, et al. Heterogeneous distribution of BRAF/NRAS mutations among Italian patients with advanced melanoma. J Transl Med. 2013;11(1):202.
Boursault L, Haddad V, Vergier B, et al. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One. 2013;8(8):e70826.
Picard M, Pham Dang N, D’Incan M, et al. Is BRAF a prognostic factor in stage III skin melanoma? A retrospective study of 72 patients after positive sentinel lymph node dissection. Br J Dermatol. 2014;171(1):108-14.
Saint-Jean M, Quéreux G, Nguyen J-M, et al. Is a single BRAF wild-type test sufficient to exclude melanoma patients from vemurafenib therapy? J Investig Dermatol. 2014;134(5):1468-70.
Sakaizawa K, Ashida A, Uchiyama A, et al. Clinical characteristics associated with BRAF, NRAS and KIT mutations in Japanese melanoma patients. J Dermatol Sci. 2015;80(1):33-7.
Eriksson H, Zebary A, Vassilaki I, et al. BRAFV600E protein expression in primary cutaneous malignant melanomas and paired metastases. JAMA Dermatol. 2015;151(4):410-6.
Nardin C, Puzenat E, Prétet JL, et al. BRAF mutation screening in melanoma: is sentinel lymph node reliable? Melanoma Res. 2015;25(4):328-34.
Wang F, Fang P, Hou D-Y, et al. Comparison of epidermal growth factor receptor mutations between primary tumors and lymph nodes in non-small cell lung cancer: a review and meta-analysis of published data. Asian Pac J Cancer Prev. 2014;15(11):4493-7.
Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507-16.
Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707-14.
Cheng L, Lopez-Beltran A, Massari F, et al. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod Pathol. 2018;31(1):24-38.