Corresponding author: Masaki Shiota, shiota@uro.med.kyushu-u.ac.jp
DOI: 10.31662/jmaj.2021-0004
Received: January 12, 2021
Accepted: January 15, 2021
Advance Publication: March 26, 2021
Published: April 15, 2021
Cite this article as:
Shiota M, Akamatsu S, Narita S, Terada N, Fujimoto N, Eto M. Genetic Polymorphisms and Pharmacotherapy for Prostate Cancer. JMA J. 2021;4(2):99-111.
The therapeutic landscape of pharmacotherapy for prostate cancer has dramatically evolved, and multiple therapeutic options have become available for prostate cancer patients. Therefore, useful biomarkers to identify suitable candidates for treatment are required to maximize the efficacy of pharmacotherapy. Genetic polymorphisms such as single-nucleotide polymorphisms (SNPs) and tandem repeats have been shown to influence the therapeutic effects of pharmacotherapy for prostate cancer patients. For example, genetic polymorphisms in the genes involved in androgen receptor signaling are reported to be associated with the therapeutic outcome of androgen-deprivation therapy as well as androgen receptor-pathway inhibitors. In addition, SNPs in genes involved in drug metabolism and efflux pumps are associated with therapeutic effects of taxane chemotherapy. Thus, genetic polymorphisms such as SNPs are promising biomarkers to realize personalized medicine. Here, we overview the current findings on the influence of genetic polymorphisms on the outcome of pharmacotherapy for prostate cancer and discuss current issues as well as future visions in this field.
Key words: androgen metabolism, androgen receptor, genetic polymorphism, pharmacotherapy, prostate cancer
Androgen-deprivation therapy (ADT) with or without first-generation anti-androgen agents has been the gold standard as primary pharmacotherapy for treatment-naïve prostate cancer (1). Recently, the therapeutic landscape of pharmacotherapy for prostate cancer patients has been greatly evolving. Second-generation anti-androgen agents such as enzalutamide, apalutamide, and darolutamide as well as the CYP17 inhibitor abiraterone have been developed for castration-resistant prostate cancer (CRPC) (2). Although these drugs were initially developed for the treatment of CRPC, enzalutamide, apalutamide, and abiraterone have expanded for use in hormone-sensitive prostate cancer (HSPC) (3). In addition, taxane chemotherapy (docetaxel and cabazitaxel) and radioisotopes (radium-223) have been applied for the treatment of CRPC, and docetaxel has been indicated for HSPC (2). Thus, multiple therapeutic options for CRPC and HSPC are available. Therefore, useful biomarkers to identify patients that are suitable candidates for these treatments are required to maximize the efficacy of pharmacotherapy.
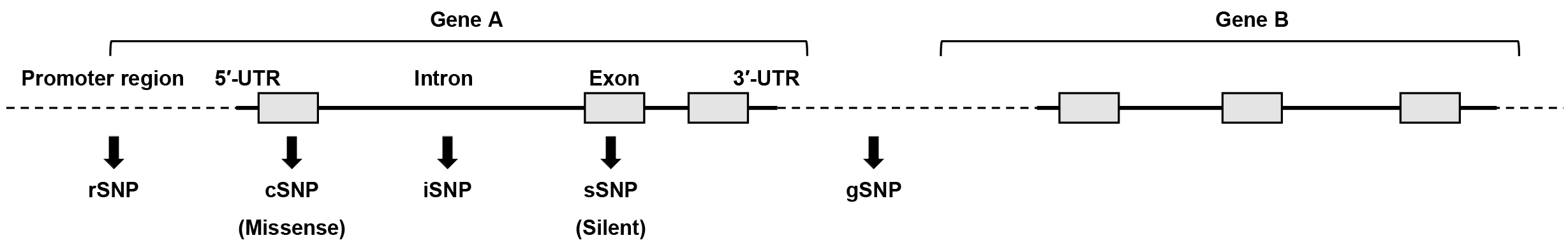
Genetic polymorphisms are considered one of the most promising biomarkers for the realization of personalized medicine (4). Genetic polymorphisms are inter-individual differences in germline DNA and defined as differences in genomic sequences between individuals that occur at a frequency of 1% or more in a population. Most genetic polymorphisms are single-nucleotide polymorphisms (SNPs), and polymorphisms are also detected in repeated sequences such as microsatellites. SNPs are observed at a frequency of ~1 in 1000 nucleotides, and more than 2 million SNPs exist in the entire human genome. SNPs are classified into the following types according to their function: regulatory SNPs (rSNPs), which are located in promoter regions; coding SNPs (cSNPs), which are located in exons and cause an amino acid substitution; silent SNPs (sSNPs), which are located in an exon but do not cause an amino acid substitution; intron SNPs (iSNPs), which are located in introns; and genome SNPs (gSNPs), which are located in intergenic regions (Figure 1). Accordingly, rSNPs and cSNPs are likely to change gene expression and protein function, which results in functional and phenotypic differences, respectively. In addition, sSNPs and iSNPs may affect expression levels of genes. Conversely, gSNPs are speculated to not play a direct functional role, but these may serve as genomic markers linked with distinct functional SNPs.
Genetic polymorphisms can cause various phenotypic differences through changes of expression and/or activity in the corresponding gene. Genetic polymorphisms are also associated not only with disease susceptibility but also with treatment outcomes. For example, a genetic polymorphism in UGT1A1 (UGT1A1*28 and UGT1A1*6), which encodes UDP-glucuronosyltransferase, decreases enzyme activity, and delays the metabolism of SN-38, the active metabolite of irinotecan, which results in a higher incidence of adverse events by irinotecan (5). A test for genetic polymorphisms in UGT1A1 has been approved in Japan for patients who will be treated with irinotecan chemotherapy. A SNP in Nudix hydrolase 15 (NUDT15), which encodes the enzyme involved in the metabolism of thiopurines, was shown to be useful in predicting adverse events of thiopurines. A test for genetic polymorphisms in NUDT15 was recently approved in Japan for patients who will be treated with thiopurines (6). Thus, the significance of testing genetic polymorphisms including SNPs in medical care has been growing.
Several genome-wide associated studies (GWASs) on prostate cancer susceptibility in large cohorts have been reported, showing the value of hundreds of SNPs with prostate cancer incidence (7), (8). In addition, various studies have reported the significance of SNPs in the outcome of pharmacotherapy for prostate cancer (9). An association of genetic background such as race and family history with the outcome of prostate cancer has been shown, which suggests that genetic factors play an important role in pharmacotherapy (10), (11). In this review, we provide an overview of the current findings on the influence of genetic polymorphisms in pharmacotherapy for prostate cancer and discuss current issues and future directions in this field.
Aberrant activation of androgen receptor (AR) signaling is one of the main causes by which prostate cancer acquires castration resistance. Therefore, polymorphisms in genes related to the AR pathway may affect the therapeutic efficacy of primary ADT through influencing AR signaling activity (9). To date, 63 SNPs in 49 genes have been reported to be associated with the outcome of primary ADT for HSPC (Table 1).
Table 1. Genetic Polymorphisms Associated with Treatment Outcomes for Hormone-Sensitive Prostate Cancer.
| Gene name | Function | rs number | Polymorphism types | Treatment | Validation | Reference |
|---|---|---|---|---|---|---|
| CYP17A1 | Androgen metabolism | rs6162 | sSNP | ADT | (12) | |
| rs743572 | rSNP | ADT | Validated | (13), (14) | ||
| CYP19A1 | Androgen metabolism | rs1870050 | iSNP | ADT | Almost validated | (12), (15), (16) |
| rs4775936 | iSNP | ADT | (17) | |||
| HSD3B1 | rs1047303 | cSNP | ADT | Validated | (18)-(22) | |
| ADT+Docetaxel | (22) | |||||
| rs1856888 | gSNP | ADT | Almost validated | (15), (23) | ||
| HSD17B2 | Androgen metabolism | rs4243229, rs7201637 | iSNP | ADT | (12) | |
| HSD17B3 | Androgen metabolism | rs2257157 | iSNP | ADT | (12) | |
| HSD17B4 | Androgen metabolism | rs7737181 | iSNP | ADT | (15) | |
| AKR1C3 | Androgen metabolism | rs12529 | cSNP | ADT | Controversial | (24), (25) |
| SRD5A2 | Androgen metabolism | rs523349 | cSNP | ADT | (26) | |
| SLCO1B3 | Androgen transporter | rs4149117 | cSNP | ADT | Validated | (27)-(29) |
| SLCO2B1 | Androgen transporter | rs1077858 | iSNP | ADT | Validated | (30), (31) |
| rs1789693 | iSNP | ADT | (30) | |||
| rs12422149 | cSNP | ADT | Almost validated | (29)-(32) | ||
| GNRH2 | Androgen synthesis | rs6051545 | cSNP | ADT | (33) | |
| SHBG | Androgen-binding protein | rs6259 | cSNP | ADT | Controversial | (34), (35) |
| AR | Steroid receptor | CAG repeat | Coding region | ADT | Almost validated | (24), (36), (37) |
| ESR1 | Steroid receptor | rs1062577 | rSNP | ADT | (12) | |
| NR3C2 | Steroid receptor | rs5522 | cSNP | ADT | (38) | |
| YB-1 | Transcription factor | rs12030724 | iSNP | ADT | Validated | (39), (40) |
| HIF1A | Transcription factor | rs11549465 | cSNP | ADT | (41) | |
| ARRDC3 | Target gene of AR | rs2939244 | rSNP | ADT | (42) | |
| FLT1 | Target gene of AR | rs9508016 | rSNP | ADT | (42) | |
| SKAP1 | Target gene of AR | rs6054145 | rSNP | ADT | (42) | |
| FBXO32 | Target gene of AR | rs7830622 | rSNP | ADT | (42) | |
| BNC2 | Target gene of ER | rs16934641 | rSNP | ADT | (43) | |
| TACC2 | Target gene of ER | rs3763763 | rSNP | ADT | (43) | |
| ALPK1 | Target gene of ER | rs2051778 | rSNP | ADT | (43) | |
| LSAMP | Target gene of NFκB | rs13088089 | rSNP | ADT | (44) | |
| CCL17 | Target gene of NFκB | rs223899 | rSNP | ADT | (44) | |
| PSMD7 | Target gene of NFκB | rs2387084 | rSNP | ADT | (44) | |
| MON1B | Target gene of NFκB | rs284924 | rSNP | ADT | (44) | |
| GSTM3 | Antioxidant | rs7483 | cSNP | ADT | Validated | (45) |
| CAT | Antioxidant | rs564250 | gSNP | ADT | (45) | |
| SLC28A3 | Nucleoside transporter | rs56350726 | cSNP | ADT | (46) | |
| LRP2 | Sterol and steroid transporter | rs6433107, rs3944004, rs830994, rs3770613, rs831003 | iSNP | ADT | (47) | |
| EGF | Growth factor | rs4444903 | rSNP | ADT | (48) | |
| IRS2 | Growth factor | rs7986346 | gSNP | ADT | (49) | |
| TGFBR2 | TGF-β signaling | rs3087465 | iSNP | ADT | (50) | |
| BMP5 | TGF-β signaling | rs317027 | gSNP | ADT | (49) | |
| IL18 | Cytokine | rs187238 | rSNP | ADT | (51) | |
| APC | Wnt signaling | rs2707765, rs497844 | iSNP | ADT | (52) | |
| BGLAP | Bone metabolism | rs1800247 | rSNP | ADT | (53) | |
| EDN1 | Vasoconstrictor | rs1800541, rs2070699 | iSNP | ADT | (54) | |
| CASP3 | Apoptosis | rs4862396 | gSNP | ADT | (49) | |
| TRMT11 | Methyltransferase | rs1268121, rs6900796 | iSNP | ADT | (55) | |
| COMT | Methyltransferase | rs4680 | cSNP | Estramustine phosphate | (56) | |
| KIF3C | miRNA target site | rs6728684 | rSNP | ADT | (57) | |
| CDON | miRNA target site | rs3737336 | rSNP | ADT | (57) | |
| IFI30 | miRNA target site | rs1045747 | rSNP | ADT | (57) | |
| PALLD | miRNA target site | rs1071738 | rSNP | ADT | (57) | |
| GABRA1 | miRNA target site | rs998754 | rSNP | ADT | (57) | |
| SYT9 | miRNA target site | rs4351800 | rSNP | ADT | (57) | |
| - | - | rs16901979, rs7931342 | gSNP | ADT | (58) | |
| ADT, androgen deprivation therapy; AR, androgen receptor; ER, estrogen receptor; NFκB, nuclear factor-κ B; SNP, single-nucleotide polymorphism; TGF, tumor growth factor | ||||||
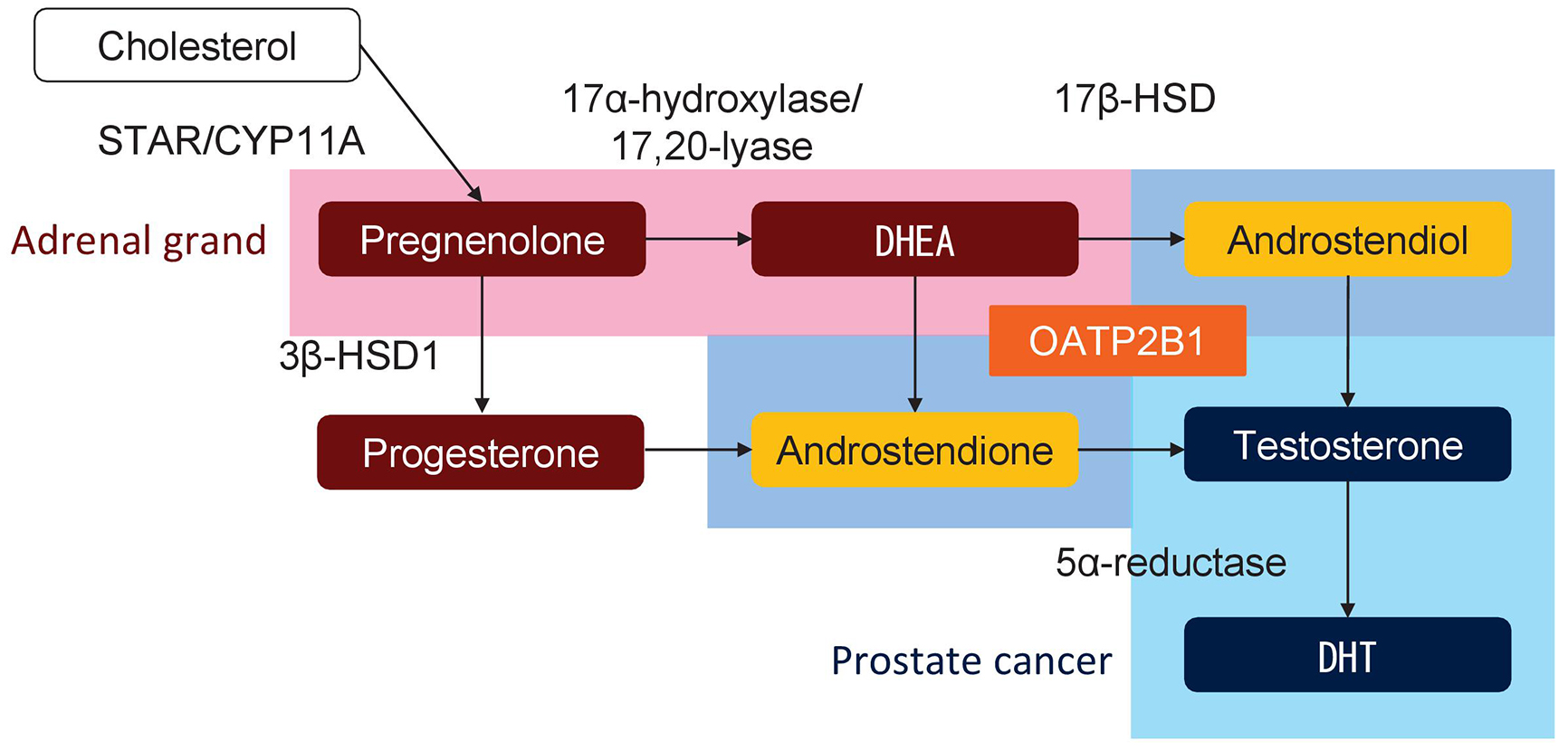
De novo androgen synthesis in prostate cancer cells is a major source of androgen under castrated condition during ADT and is shown to play an important role in the progression to CRPC (59). Multiple studies have indicated the association of SNPs in genes involved in androgen metabolism, including CYP17A1, CYP19A1, HSD3B1, HSD17B2, HSD17B3, HSD17B4, AKR1C3, and SRD5A2, with the outcome of ADT (Figure 2). For example, a cSNP (rs1047303, 1245A>C, N367T) in HSD3B1, which encodes 3β-hydroxysteroid dehydrogenase 1 (3β-HSD1), results in a variant of 3β-HSD1 with high activity, and the prognosis of carriers of this variant is poor (18), (19), (20), (21), (22), (60). The prognostic impact of the cSNP (rs1047303) in HSD3B1 in the United States was validated in an Asian cohort (21), although variant carriers were rare in Asian patients (~15%) compared with Caucasian patients (~50%) (Table 2). The prognostic impact of the cSNP (rs1047303) in HSD3B1 was validated in primary ADT plus docetaxel for HSPC (22). In addition, the prognostic difference by another SNP (rs1856888) in HSD3B1 was also indicated. Ross et al. initially reported that the variant G allele in rs1856888 was associated with a low risk of disease progression among men in the United States (15); however, a recent study from the United States showed poor prognosis in patients carrying the variant G allele in rs1856888 (23). Because of the strong linkage disequilibrium between the SNPs (rs1047303 and rs1856888) in HSD3B1 (23), the variant allele in the SNPs (rs1047303 and rs1856888) in HSD3B1 is likely to be associated with poor prognosis in patients treated with primary ADT. In a study on an iSNP (rs1870050) in CYP19A1, Ross et al. reported that the variant C allele in rs1870050 was associated with a high risk of disease progression among men in the United States (15). However, two recent studies showed a low risk of progression and better prognosis among Asian men with the variant C allele in rs1870050 (12), (16). In addition, the prognostic significance of an rSNP (rs743572) in the 5′ untranslated region of CYP17A1 has been shown (13), (14).
Table 2. Outcome and Frequencies of the rs1047303 Variant Allele of HSD3B1.
| Outcome | Variant carrier | Number | Frequency carrying a variant allele | Reference |
|---|---|---|---|---|
| Prostate cancer susceptibility | High | 626 | 48% (AC/CC, US) | (62) |
| Hereditary prostate cancer susceptibility | High | 98 | 53% (AC/CC, US) | (63) |
| Prognosis in primary ADT | Poor | 118/137/118 | 51% (AC/CC, US) | (18) |
| Prognosis in primary ADT | Poor | 102 | 53% (AC/CC, US) | (19) |
| Prognosis in primary ADT | Poor | 218 | 54% (AC/CC, US) | (20) |
| Prognosis in Abiraterone | Insignificant | 76 | 45% (AC/CC, US) | (64) |
| Progression in primary ADT or ADT+Docetaxel | Poor in low volume | 475 | 53% (AC/CC, US) | (22) |
| Prognosis in Ezalutamide or Abiraterone | Poor | 266 | 8% (CC, US/UK) | (65) |
| Prognosis in Ezalutamide or Abiraterone | Poor | 547 | 15% (CC, Canada/Europe) | (66) |
| Prognosis in primary ADT | Insignificant | 103 | 18% (AC/CC, China) | (67) |
| Prognosis in primary ADT | Poor | 104 | 9% (AC/CC, Japan) | (21) |
| Prognosis in Abiraterone | Favorable | 99 | 14% (AC/CC, Japan) | (21) |
| ADT, androgen deprivation therapy; UK, United Kingdom; US, United States | ||||
In addition to enzymes for androgen metabolism, the pump for androgens such as dehydroepiandrosterone (DHEA) and testosterone also plays a key role in the development of CRPC (29). SNPs in SLCO1B3 and SLCO2B1 genes, which encode proteins responsible for the import of testosterone and DHEA, respectively, were reported to be associated with the prognosis of patients treated with primary ADT (27), (28), (29), (30), (31), (32). Higher testosterone uptake in patients with the variant allele of the cSNP (rs4149117, 334G>T, A112S) in SLCO1B3 was shown, and a causal variant of SLCO1B3 was reported to be associated with poor prognosis (27), (28), (29). Several studies demonstrated that the cSNP (rs12422149, 935G>A, R312Q) in SLCO2B1 resulted in higher activity of DHEA-sulfate uptake and the wild-type allele in SLCO2B1 (rs12422149) was associated with early recurrence and poor prognosis after primary ADT (29), (30), (31), (32). Another SNP (rs1077858) in SLCO2B1 was also associated with prognosis (30), (31). Thus, SNPs in the genes involved in androgen metabolism and uptake in prostate cancer cells play a key role in the progression of prostate cancer through persistent androgen synthesis in prostate cancer under castrated condition (Figure 3).
SNPs in other molecules related to the AR pathway were also shown to have prognostic impact after primary ADT. For example, the CAG repeat in AR correlated with prognosis, although null results were also reported (24), (36), (37), (61). In addition, the iSNP (rs12030724) in YB-1 that regulates YB-1 expression, which results in AR and AR variant expression, was also associated with the prognosis of Japanese men with advanced prostate cancer treated with primary ADT (39), (40). The cSNP (rs7483, I224V) in GSTM3, which encodes an antioxidant enzyme, was also reported to be prognostic in Japanese patients with nonmetastatic and advanced prostate cancer treated with primary ADT (45).
Novel ARPIs such as enzalutamide, apalutamide darolutamide, and abiraterone have been demonstrate to improve survival in patients with CRPC (2). Because abiraterone is taken up into cells by OATP2B1, which is encoded by SLCO2B1, and then metabolized by 3β-HSD and 5α-reductase, the therapeutic effect of abiraterone treatment may depend on the activities of the molecules involved in androgen metabolism and uptake (Figure 2) (73), (78). Recent reports showed that SNPs in genes involved in androgen metabolism and transport such as CYP17A1, HSD3B1, SRD5A2, and SLCO2B1 correlate with the outcome of abiraterone treatment (Table 3). An rSNP (rs2486758, -362T>C) in CYP17A1 was associated with prognosis after abiraterone treatment (68), (69). In addition, variant carriers of the cSNP (rs1047303) in HSD3B1 showed poor prognosis after treatment with ARPI (65), (66). The prognostic impact of the cSNP (rs1047303) in HSD3B1 for both primary ADT for HSPC as well as ARPIs for CRPC may be because of hyperactive androgen synthesis in variant carriers. The variant allele in HSD3B1 is expected to lead to increased conversion from abiraterone to the more potent delta-4-abiraterone (78). Accordingly, the cSNP (rs1047303) in HSD3B1 was shown to be associated with comparable or better treatment efficacy of abiraterone (21), (64).
Table 3. Genetic Polymorphisms Associated with the Prognosis of Patients with Castration-Resistant Prostate Cancer Treated with Androgen Receptor-Pathway Inhibitors.
| Gene name | Function | rs number | Polymorphism types | Treatment | Validation | Reference |
|---|---|---|---|---|---|---|
| CYP17A1 | Androgen metabolism | rs2486758 | rSNP | Abiraterone | Validated | (68), (69) |
| rs10883783 | iSNP | Abiraterone | (70) | |||
| HSD3B1 | Androgen metabolism | rs1047303 | cSNP | Abiraterone | (21) | |
| Abiraterone or Enzalutamide | Validated | (65), (66) | ||||
| SRD5A2 | Androgen metabolism | rs523349 | cSNP | Abiraterone | (71) | |
| SLCO2B1 | Androgen transporter | rs1077858, rs1789693, rs34550074 | iSNP, iSNP, cSNP | Abiraterone | (72) | |
| rs12422149 | cSNP | Abiraterone | (73) | |||
| YB-1 | Androgen receptor regulator | rs10493112 | iSNP | Abiraterone | (74) | |
| CYB5A | CYP17A1 activity regulator | rs1790834 | iSNP | Abiraterone | (75) | |
| TSPYL1 | CYP17A1 and CYP3A4 regulator | rs3828743 | cSNP | Abiraterone | (76) | |
| SULT1E1 | Estrogen metabolism | Group 1 (rs3775777, rs4149534, rs10019305) | iSNP | Abiraterone | (77) | |
| Group 2 (rs3775770, rs4149527, rs3775768) | ||||||
| SNP, single-nucleotide polymorphism | ||||||
Interestingly, several genes are overlapping in association with the prognosis between primary ADT and ARPIs, which both target the AR pathway (Table 4). SNPs in CYP17A1 and YB-1 are associated with the outcome of primary ADT and ARPIs, although the SNPs in each gene are different. Furthermore, the cSNP (rs1047303) in HSD3B1, cSNP (rs523349) in SRD5A2, and SNPs (rs1077858, rs1789693, and rs12422149) in SLCO2B1 were shown to be common prognosticators in both primary ADT for HSPC and ARPIs for CRPC. The prognostic and antitumor impacts of the cSNP (rs523349) in SRD5A2 and SNPs (rs1077858, rs1789693, and rs12422149) in SLCO2B1 were consistent between primary ADT and abiraterone. Intriguingly, a variant allele in HSD3B1 (rs1047303) was differentially associated with prognosis in patients treated with abiraterone and other therapies. These findings suggest that HSD3B1 (rs1047303) may be a promising marker to select appropriate combination therapy with ADT. Further studies on the prognostic impact of these SNPs will be important to evaluate candidates for personalized medicine.
Table 4. Genetic Polymorphisms Associated with the Outcomes of Multiple Treatments.
| Gene name | rs number | Treatment regimen | Risk allele | Outcome | Reference |
|---|---|---|---|---|---|
| CYP17A1 | rs6162 | ADT | G | OS | (12) |
| rs743572 | ADT | A | OS | (13) | |
| ADT | A | PFS | (14) | ||
| rs2486758 | Abiraterone | C | PFS | (68) | |
| Abiraterone | C | PFS | (69) | ||
| rs10883783 | Abiraterone | A | PFS | (70) | |
| HSD3B1 | rs1047303 | ADT | C | PFS, MFS, OS | (18) |
| ADT | C | PFS | (19) | ||
| ADT | C | MFS | (20) | ||
| ADT | C | PFS | (21) | ||
| ADT | C | PFS, OS | (22) | ||
| ADT+Docetaxel | C | PFS, OS | (22) | ||
| Abiraterone | Null | PFS | (64) | ||
| Abiraterone | A | PFS, OS | (21) | ||
| Abiraterone/Enzalutamide | C | OS | (65) | ||
| Abiraterone/Enzalutamide | C | PFS | (66) | ||
| rs1856888 | ADT | A | PFS | (15) | |
| ADT | G | OS | (23) | ||
| SRD5A2 | rs523349 | ADT | G | PFS, OS | (26) |
| Abiraterone | G | PFS | (71) | ||
| SLCO1B3 | rs4149117 | ADT | T | OS | (27) |
| T | PFS | (28) | |||
| T | CSS | (29) | |||
| Cabazitaxel | Null | OS | (79) | ||
| SLCO2B1 | rs1077858 | ADT | G | PFS | (30) |
| ADT | G | OS | (31) | ||
| Abiraterone | G | MRD | (72) | ||
| rs1789693 | ADT | T | PFS | (30) | |
| Abiraterone | T | MRD | (72) | ||
| rs34550074 | Abiraterone | T | MRD | (72) | |
| rs12422149 | ADT | A | CSS | (29) | |
| ADT | G | PFS | (30) | ||
| ADT | G | PFS | (32) | ||
| ADT | G | PFS | (31) | ||
| Abiraterone | G | PFS | (73) | ||
| YB-1 | rs10493112 | Abiraterone | A | PFS | (74) |
| rs12030724 | ADT | A | PFS | (39) | |
| ADT | A | PFS, OS | (40) | ||
| ESR1 | rs1062577 | ADT | A | OS | (12) |
| rs2234693 | Docetaxel | C | PFS | (80) | |
| rs9340799 | Docetaxel+Thalidomide | G | PFS | (80) | |
| ADT, androgen deprivation therapy; MRD, minimal residual disease; OS, overall survival; PFS, progression-free survival | |||||
The taxane docetaxel is widely used not only for prostate cancer but also for various cancers such as lung, uterine, and ovarian cancers. Many reports have demonstrated the relationship between genetic polymorphisms and the efficacy and adverse events of docetaxel therapy. Previous studies, including several in prostate cancer, reported associations between drug transport genes (ABCB1, ABCC2, ABCG1, ABCG2, SLCO1B3) or drug metabolism genes (CYP1B1, CYP2C8, CYP3A4, CYP3A5) with therapeutic efficacy or adverse events (Figure 2) (81). As shown in Table 5, the cSNP (rs1056836, 4326C>G, L432V) in CYP1B1 was associated with poor response and prognosis (82), (83). In addition, SNPs in estrogen receptor 1 (ESR1) were also associated with treatment efficacy in prostate cancer (80). SNPs in ESR1 were reported to be associated with the outcome of primary ADT and taxane chemotherapy although the position of SNPs in ESR1 is different (Table 4). Then, SNPs in ESR1 may serve as a predictive marker for taxane chemotherapy. OATP1B3, which is encoded by SLCO1B3, plays a role in taxane uptake into cells and is involved in taxane resistance in prostate cancer cells. SNPs in SLCO1B3 may be associated with the treatment efficacy of taxane (88). However, a recent study showed comparable prognosis after cabazitaxel for CRPC between genotypes in SLCO1B3 (rs4149117) (79). Because prognostic impact in primary ADT has been shown(27), (28), (29), the cSNP (rs4149117) in SLCO3B1 may serve as a predictive marker in pharmacotherapy for prostate cancer.
Table 5. Genetic Polymorphisms Associated with the Outcome of Taxane Treatment for Castration-Resistant Prostate Cancer.
| Gene name | Function | rs number | Polymorphism types | Treatment | Validation | Reference |
|---|---|---|---|---|---|---|
| CYP1B1 | Drug metabolizing enzyme | rs1056836 | cSNP | Docetaxel | Validated | (82), (83) |
| ABCB1 | Drug excretion pump | rs1128503, rs2032582, rs1045642 | cSNP | Docetaxel+Thalidomide | (84) | |
| ABCB11 | Drug excretion pump | rs7602171 | iSNP | Docetaxel+Thalidomide | (85) | |
| ABCG2 | Drug excretion pump | rs2231142 | cSNP | Docetaxel+Vinorelbine/Estramustine phosphate | (86) | |
| ESR1 | Steroid receptor | rs2234693, rs9340799 | iSNP | Docetaxel+Thalidomide | (80) | |
| GSTP1 | Antioxidant | rs1138272 | cSNP | Docetaxel+Thalidomide | (85) | |
| SLC5A6 | Transporter | rs1395 | cSNP | Docetaxel+Thalidomide | (85) | |
| VEGFA | Angiogenesis | rs1570360 | rSNP | Docetaxel, Celecoxib+Cyclophosphamide | (87) | |
| SNP, single-nucleotide polymorphism | ||||||
The associations between multiple SNPs and therapeutic effects of pharmacotherapy for prostate cancer have been reported, as described above. However, to date, no genetic marker has been clinically utilized in pharmacotherapy for prostate cancer, which suggests potential issues as described in the following. While some SNPs have been reproducible in validation studies, others have not yielded consistent results across studies (Table 1, 3, 5). This may be because of racial differences in the frequency of genetic polymorphisms and linkage disequilibrium (a phenomenon in which there is a correlation between genetic polymorphisms in a population). To resolve this issue, multiple studies with large populations and meta-analysis studies are required. In addition, advances in technology such as artificial intelligence may serve as a breakthrough method to resolve the complex linkage disequilibrium among individuals.
Another problem is that the data in most study cohorts were retrospectively collected in daily practice. A daily clinical follow-up generally shows deviations from the strict follow-up schedule in a clinical trial. To improve the quality of data, collecting clinical data using a strict protocol is desirable to obtain more robust findings. In addition, most studies to date have focused on target genetic polymorphisms of individual genes. Because this method may miss useful SNPs, comprehensive methods such as GWAS are required. In addition, a single marker may be not enough for accurate predictive ability, and this may be overcome by using multiple SNPs. GWASs indicated that a single SNP generally provides only a modest (odds ratio, 1.1-1.5) increased susceptibility risk of prostate cancer, where polygenic risk score (PGS) using multiple risk SNPs was developed and validated (89), (90). Therefore, the PGS approach would be useful to increase diagnostic ability.
Furthermore, the genes of SNPs associated with therapeutic outcome can be the cause of treatment resistance. Therefore, these genes are promising targets to overcome treatment resistance. Genes involved in androgen metabolism such as CYP17A1, HSD3B1, AKR1C3, and SRD5A2 have been candidate targets for drug discovery and drug development, and the SNPs may be crucial in therapeutic efficacy (Table 6).
Table 6. Druggable Targets in Androgen Metabolism and Their Inhibitors.
| Target enzyme | Inhibitor | Developmental status | |
|---|---|---|---|
| CYP17 | Abiraterone | Approved | |
| Orteronel (TAK-700) | Phase III (terminated) | ||
| Galeterone | Phase II (terminated) | ||
| 3β-HSD | Trilostane | Phase II (terminated)/on market for Cushing's syndrome | |
| AKR1C3 | Indometacin | Phase II/on market as NSAIDs | |
| N-(indolylcarbonyl)-piperidines | Phase I | ||
| 5α-reductase (types I and II) | Dutasteride | Phase II (terminated)/on market for benign prostatic hyperplasia | |
| 5α-reductase (type II) | Finasteride | On market for androgenetic alopecia | |
| NSAID, non-steroidal anti-inflammatory drug | |||
Here, we summarized the known associations between genetic polymorphisms and the outcomes of pharmacotherapy in prostate cancer patients. Recently, multiple novel therapeutic options for HSPC have emerged, and the stratification of suitable patients for each option will be required. Genetic biomarkers such as SNPs will be beneficial for stratifying patients and for estimating the treatment́ response of an individual patient. The combination of genetic biomarkers with traditional clinicopathological parameters could improve the prognostication and the choice of the most appropriate treatment for each patient, which will be helpful in clinical decision making. Thus, personalized medicine using genetic biomarkers is expected to be realized in pharmacotherapy for prostate cancer. However, unresolved issues remain, such as inconsistent results among studies as well as the current lack of GWAS and PGS approaches, and these issues should be addressed in future research.
This article is based on the study, which received the Medical Research Encouragement Prize of The Japan Medical Association in 2020.
Masaki Shiota received honoraria from Janssen Pharmaceutical Company, Astellas Pharma, and Sanofi; Shusuke Akamatsu received grant/research support from Astellas Pharma; Shintaro Narita received honoraria from Janssen Pharmaceutical Company; Naohiro Fujimoto received honoraria from Janssen Pharmaceutical Company and Astellas Pharma and grant/research support from Astellas Pharma and Sanofi; Masatoshi Eto received honoraria from Janssen Pharmaceutical Company and grant/research support from Astellas Pharma and Sanofi.
This work was supported by Takeda Science Foundation and Japanese Urological Association to Masaki Shiota.
We thank Gabrielle White Wolf, PhD, from Edanz Group (https://en-author-services.edanz.com/ac) for editing a draft of this manuscript.
Shiota M, Eto M. Current status of primary pharmacotherapy and future perspectives toward upfront therapy for metastatic hormone-sensitive prostate cancer. Int J Urol. 2016;23(5):360-9.
Moussa M, Papatsoris A, Abou Chakra M, et al. Pharmacotherapeutic strategies for castrate-resistant prostate cancer. Expert Opin Pharmacother. 2020;21(12):1431-48.
Sathianathen NJ, Koschel S, Thangasamy IA, et al. Indirect comparisons of efficacy between combination approaches in metastatic hormone-sensitive prostate cancer: a systematic review and network meta-analysis. Eur Urol. 2020;77(3):365-72.
Choudhury AD, Eeles R, Freedland SJ, et al. The role of genetic markers in the management of prostate cancer. Eur Urol. 2012;62(4):577-87.
Hulshof EC, Deenen MJ, Guchelaar HJ, et al. Pre-therapeutic UGT1A1 genotyping to reduce the risk of irinotecan-induced severe toxicity: ready for prime time. Eur J Cancer. 2020;141:9-20.
Kakuta Y, Kawai Y, Okamoto D, et al. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: a multicenter study. J Gastroenterol. 2018;53(9):1065-78.
Akamatsu S, Takata R, Haiman CA, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44(4):426-9.
Benafif S, Kote-Jarai Z, Eeles RA; PRACTICAL Consortium. A review of prostate cancer genome-wide association studies (GWAS). Cancer Epidemiol Biomarkers Prev. 2018;27(8):845-57.
Fujimoto N, Shiota M, Tomisaki I, et al. Gene polymorphism-related individual and interracial differences in the outcomes of androgen deprivation therapy for prostate cancer. Clin Genitourin Cancer. 2017;15(3):337-42.
Fukagai T, Namiki TS, Carlile RG, et al. Comparison of the clinical outcome after hormonal therapy for prostate cancer between Japanese and Caucasian men. BJU Int. 2006;97(6):1190-3.
Hemminki K, Ji J, Försti A, et al. Concordance of survival in family members with prostate cancer. J Clin Oncol. 2008;26(10):1705-9.
Lévesque É, Huang SP, Audet-Walsh É, et al. Molecular markers in key steroidogenic pathways, circulating steroid levels, and prostate cancer progression. Clin Cancer Res. 2013;19(3):699-709.
Hamada A, Danesi R, Price DK, et al. Association of a CYP17 polymorphism with overall survival in Caucasian patients with androgen-independent prostate cancer. Urology. 2007;70(2):217-20.
Yamada T, Nakayama M, Shimizu T, et al. Genetic polymorphisms of CYP17A1 in steroidogenesis pathway are associated with risk of progression to castration-resistant prostate cancer in Japanese men receiving androgen deprivation therapy. Int J Clin Oncol. 2013;18(4):711-7.
Ross RW, Oh WK, Xie W, et al. Inherited variation in the androgen pathway is associated with the efficacy of androgen-deprivation therapy in men with prostate cancer. J Clin Oncol. 2008;26(6):842-7.
Shiota M, Fujimoto N, Tsukahara S, et al. The impact of genetic polymorphism on CYP19A1 in androgen-deprivation therapy among Japanese men. Cancer Chemother Pharmacol. 2019;83(5):933-8.
Kanda S, Tsuchiya N, Narita S, et al. Effects of functional genetic polymorphisms in the CYP19A1 gene on prostate cancer risk and survival. Int J Cancer. 2015;136(1):74-82.
Hearn JWD, AbuAli G, Reichard CA, et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 2016;17(10):1435-44.
Agarwal N, Hahn AW, Gill DM, et al. Independent validation of effect of HSD3B1 genotype on response to androgen-deprivation therapy in prostate cancer. JAMA Oncol. 2017;3(6):856-7.
Hearn JWD, Xie W, Nakabayashi M, et al. Association of HSD3B1 genotype with response to androgen-deprivation therapy for biochemical recurrence after radiotherapy for localized prostate cancer. JAMA Oncol. 2018;4(4):558-62.
Shiota M, Narita S, Akamatsu S, et al. Association of missense polymorphism in HSD3B1 with outcomes among men with prostate cancer treated with androgen-deprivation therapy or abiraterone. JAMA Netw Open. 2019;2(2):e190115.
Hearn JWD, Sweeney CJ, Almassi N, et al. HSD3B1 genotype and clinical outcomes in metastatic castration-sensitive prostate cancer. JAMA Oncol. 2020;6(4):e196496.
Chen WS, Feng EL, Aggarwal R, et al. Germline polymorphisms associated with impaired survival outcomes and somatic tumor alterations in advanced prostate cancer. Prostate Cancer Prostatic Dis. 2020;23(2):316-23.
Yu CC, Huang SP, Lee YC, et al. Molecular markers in sex hormone pathway genes associated with the efficacy of androgen-deprivation therapy for prostate cancer. PLoS One. 2013;8(1):e54627.
Shiota M, Endo S, Fujimoto N, et al. Polymorphisms in androgen metabolism genes with serum testosterone levels and prognosis in androgen-deprivation therapy. Urol Oncol. 2020;38(11):849.e11-8.
Shiota M, Fujimoto N, Yokomizo A, et al. SRD5A gene polymorphism in Japanese men predicts prognosis of metastatic prostate cancer with androgen-deprivation therapy. Eur J Cancer. 2015;51(14):1962-9.
Hamada A, Sissung T, Price DK, et al. Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res. 2008;14(11):3312-8.
Sharifi N, Hamada A, Sissung T, et al. A polymorphism in a transporter of testosterone is a determinant of androgen independence in prostate cancer. BJU Int. 2008;102(5):617-21.
Wright JL, Kwon EM, Ostrander EA, et al. Expression of SLCO transport genes in castration-resistant prostate cancer and impact of genetic variation in SLCO1B3 and SLCO2B1 on prostate cancer outcomes. Cancer Epidemiol Biomarkers Prev. 2011;20(4):619-27.
Yang M, Xie W, Mostaghel E, et al. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J Clin Oncol. 2011;29(18):2565-73.
Wang X, Harshman LC, Xie W, et al. Association of SLCO2B1 genotypes with time to progression and overall survival in patients receiving androgen-deprivation therapy for prostate cancer. J Clin Oncol. 2016;34(4):352-9.
Fujimoto N, Kubo T, Inatomi H, et al. Polymorphisms of the androgen transporting gene SLCO2B1 may influence the castration resistance of prostate cancer and the racial differences in response to androgen deprivation. Prostate Cancer Prostatic Dis. 2013;16(4):336-40.
Shiota M, Fujimoto N, Takeuchi A, et al. The association of polymorphisms in the gene encoding gonadotropin-releasing hormone with serum testosterone level during androgen deprivation therapy and prognosis of metastatic prostate cancer. J Urol. 2018;199(3):734-40.
Monteiro C, Sousa MV, Ribeiro R, et al. Genetic variants in AR and SHBG and resistance to hormonal castration in prostate cancer. Med Oncol. 2013;30(1):490.
Shiota M, Fujimoto N, Tsukahara S, et al. Genetic polymorphism in sex hormone-binding globulin with a prognosis of androgen deprivation therapy in metastatic prostate cancer among Japanese men. Clin Genitourin Cancer. 2019;17(3):e387-93.
Bratt O, Borg A, Kristoffersson U, et al. CAG repeat length in the androgen receptor gene is related to age at diagnosis of prostate cancer and response to endocrine therapy, but not to prostate cancer risk. Br J Cancer. 1999;81(4):672-6.
Shimbo M, Suzuki H, Kamiya N, et al. CAG polymorphic repeat length in androgen receptor gene combined with pretreatment serum testosterone level as prognostic factor in patients with metastatic prostate cancer. Eur Urol. 2005;47(4):557-63.
Shiota M, Fujimoto N, Imada K, et al. Prognostic impact of genetic polymorphism in mineralocorticoid receptor and comorbidity with hypertension in androgen-deprivation therapy. Front Oncol. 2018;8:635.
Shiota M, Fujimoto N, Imada K, et al. Potential role for YB-1 in castration-resistant prostate cancer and resistance to enzalutamide through the androgen receptor V7. J Natl Cancer Inst. 2016;108(7):djw005.
Shiota M, Narita S, Habuchi T, et al. Validated prognostic significance of YB-1 genetic variation in metastatic prostate cancer. Pharmacogenomics J. 2021;21(1):102-5.
Fraga A, Ribeiro R, Príncipe P, et al. The HIF1A functional genetic polymorphism at locus +1772 associates with progression to metastatic prostate cancer and refractoriness to hormonal castration. Eur J Cancer. 2014;50(2):359-65.
Huang CN, Huang SP, Pao JB, et al. Genetic polymorphisms in androgen receptor-binding sites predict survival in prostate cancer patients receiving androgen-deprivation therapy. Ann Oncol. 2012;23(3):707-13.
Huang CN, Huang SP, Pao JB, et al. Genetic polymorphisms in oestrogen receptor-binding sites affect clinical outcomes in patients with prostate cancer receiving androgen-deprivation therapy. J Intern Med. 2012;271(5):499-509.
Huang SP, Lin VC, Lee YC, et al. Genetic variants in nuclear factor-kappa B binding sites are associated with clinical outcomes in prostate cancer patients. Eur J Cancer. 2013;49(17):3729-37.
Shiota M, Fujimoto N, Itsumi M, et al. Gene polymorphisms in antioxidant enzymes correlate with the efficacy of androgen-deprivation therapy for prostate cancer with implications of oxidative stress. Ann Oncol. 2017;28(3):569-75.
Jo JK, Oh JJ, Kim YT, et al. A genetic variant in SLC28A3, rs56350726, is associated with progression to castration-resistant prostate cancer in a Korean population with metastatic prostate cancer. Oncotarget. 2017;8(57):96893-902.
Holt SK, Karyadi DM, Kwon EM, et al. Association of megalin genetic polymorphisms with prostate cancer risk and prognosis. Clin Cancer Res. 2008;14(12):3823-31.
Teixeira AL, Ribeiro R, Cardoso D, et al. Genetic polymorphism in EGF is associated with prostate cancer aggressiveness and progression-free interval in androgen blockade-treated patients. Clin Cancer Res. 2008;14(11):3367-71.
Huang SP, Bao BY, Hour TC, et al. Genetic variants in CASP3, BMP5, and IRS2 genes may influence survival in prostate cancer patients receiving androgen-deprivation therapy. PLoS One. 2012;7(7):e41219.
Teixeira AL, Gomes M, Nogueira A, et al. Improvement of a predictive model of castration-resistant prostate cancer: functional genetic variants in TGFβ1 signaling pathway modulation. PLoS One. 2013;8(8):e72419.
Liu JM, Liu JN, Wei MT, et al. Effect of IL-18 gene promoter polymorphisms on prostate cancer occurrence and prognosis in Han Chinese population. Genet Mol Res. 2013;12(1):820-9.
Geng JH, Lin VC, Yu CC, et al. Inherited variants in Wnt pathway genes influence outcomes of prostate cancer patients receiving androgen deprivation therapy. Int J Mol Sci. 2016;17(12):1970.
Wu HC, Lin CC, Chen WC, et al. Osteocalcin gene HindIII C/T polymorphism is a biomarker for prostate cancer and responsiveness to hormone therapy. Eur Urol. 2003;43(2):197-200.
Xu D, Wang X, Lou Y. Association of endothelin-1 gene single-nucleotide polymorphisms and haplotypes with risk of hormone refractory prostate cancer. Pharmazie. 2017;72(2):103-6.
Kohli M, Riska SM, Mahoney DW, et al. Germline predictors of androgen deprivation therapy response in advanced prostate cancer. Mayo Clin Proc. 2012;87(3):240-6.
Suzuki M, Mamun MR, Hara K, et al. The Val158Met polymorphism of the catechol-O-methyltransferase gene is associated with the PSA-progression-free survival in prostate cancer patients treated with estramustine phosphate. Eur Urol. 2005;48(5):752-9.
Bao BY, Pao JB, Huang CN, et al. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin Cancer Res. 2011;17(4):928-36.
Bao BY, Pao JB, Huang CN, et al. Significant associations of prostate cancer susceptibility variants with survival in patients treated with androgen-deprivation therapy. Int J Cancer. 2012;130(4):876-84.
Feng Q, He B. Androgen receptor signaling in the development of castration-resistant prostate cancer. Front Oncol. 2019;9:858.
Chang KH, Li R, Kuri B, et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell. 2013;154(5):1074-84.
Misra D, Xie W, Regan MM, et al. Germline CAG repeat length of the androgen receptor and time to progression in patients with prostate cancer treated with androgen deprivation therapy. BJU Int. 2011;108(7):1086-91.
Chang BL, Zheng SL, Hawkins GA, et al. Joint effect of HSD3B1 and HSD3B2 genes is associated with hereditary and sporadic prostate cancer susceptibility. Cancer Res. 2002;62(6):1784-9.
Park JY, Tanner JP, Sellers TA, et al. Association between polymorphisms in HSD3B1 and UGT2B17 and prostate cancer risk. Urology. 2007;70(2):374-9.
Hahn AW, Gill DM, Nussenzveig RH, et al. Germline variant in HSD3B1 (1245 A > C) and response to abiraterone acetate plus prednisone in men with new-onset metastatic castration-resistant prostate cancer. Clin Genitourin Cancer. 2018;16(4):288-92.
Lu C, Terbuch A, Dolling D, et al. Treatment with abiraterone and enzalutamide does not overcome poor outcome from metastatic castration-resistant prostate cancer in men with the germline homozygous HSD3B1 c.1245C genotype. Ann Oncol. 2020;31(9):1178-85.
Khalaf DJ, Aragón IM, Annala M, et al. HSD3B1 (1245A>C) germline variant and clinical outcomes in metastatic castration-resistant prostate cancer patients treated with abiraterone and enzalutamide: results from two prospective studies. Ann Oncol. 2020;31(9):1186-97.
Wu G, Huang S, Nastiuk KL, et al. Variant allele of HSD3B1 increases progression to castration-resistant prostate cancer. Prostate. 2015;75(7):777-82.
Binder M, Zhang BY, Hillman DW, et al. Common genetic variation in CYP17A1 and response to abiraterone acetate in patients with metastatic castration-resistant prostate cancer. Int J Mol Sci. 2016;17(7):1097.
Crucitta S, Del Re M, Paolieri F, et al. CYP17A1 polymorphism c.-362T>C predicts clinical outcome in metastatic castration-resistance prostate cancer patients treated with abiraterone. Cancer Chemother Pharmacol. 2020;86(4):527-33.
Salvi S, Casadio V, Burgio SL, et al. CYP17A1 polymorphisms and clinical outcome of castration-resistant prostate cancer patients treated with abiraterone. Int J Biol Markers. 2016;31(3):e264-9.
Shiota M, Akamatsu S, Narita S, et al. The association between missense polymorphisms in SRD5A2 and HSD3B1 and treatment failure with abiraterone for castration-resistant prostate cancer. Pharmacogenomics J. Forthcoming 2021.
Mostaghel EA, Cho E, Zhang A, et al. Association of tissue abiraterone levels and SLCO genotype with intraprostatic steroids and pathologic response in men with high-risk localized prostate cancer. Clin Cancer Res. 2017;23(16):4592-601.
Hahn AW, Gill DM, Poole A, et al. Germline variant in SLCO2B1 and response to abiraterone acetate plus prednisone (AA) in new-onset metastatic castration-resistant prostate cancer (mCRPC). Mol Cancer Ther. 2019;18(3):726-9.
Afonso A, Silva J, Lopes AR, et al. YB-1 variant and androgen receptor axis-targeted agents in metastatic castration-resistant prostate cancer patients. Pharmacogenomics. 2020;21(13):919-28.
Wu X, Xu QJ, Chen PZ, et al. Association between CYP17A1, CYB5A polymorphisms and efficacy of abiraterone acetate/prednisone treatment in castration-resistant prostate cancer patients. Pharmgenomics Pers Med. 2020;13:181-8.
Qin S, Liu D, Kohli M, et al. TSPYL family regulates CYP17A1 and CYP3A4 expression: potential mechanism contributing to abiraterone response in metastatic castration-resistant prostate cancer. Clin Pharmacol Ther. 2018;104(1):201-10.
Agarwal N, Alex AB, Farnham JM, et al. Inherited variants in SULT1E1 and response to abiraterone acetate by men with metastatic castration refractory prostate cancer. J Urol. 2016;196(4):1112-6.
Li Z, Bishop AC, Alyamani M, et al. Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature. 2015;523(7560):347-51.
Belderbos BPS, de With M, Singh RK, et al. The influence of single-nucleotide polymorphisms on overall survival and toxicity in cabazitaxel-treated patients with metastatic castration-resistant prostate cancer. Cancer Chemother Pharmacol. 2020;85(3):547-53.
Sissung TM, Danesi R, Kirkland CT, et al. Estrogen receptor α and aromatase polymorphisms affect risk, prognosis, and therapeutic outcome in men with castration-resistant prostate cancer treated with docetaxel-based therapy. J Clin Endocrinol Metab. 2011;96(2):E368-72.
Frederiks CN, Lam SW, Guchelaar HJ, et al. Genetic polymorphisms and paclitaxel- or docetaxel-induced toxicities: a systematic review. Cancer Treat Rev. 2015;41(10):935-50.
Sissung TM, Danesi R, Price DK, et al. Association of the CYP1B1*3 allele with survival in patients with prostate cancer receiving docetaxel. Mol Cancer Ther. 2008;7(1):19-26.
Pastina I, Giovannetti E, Chioni A, et al. Cytochrome 450 1B1 (CYP1B1) polymorphisms associated with response to docetaxel in castration-resistant prostate cancer (CRPC) patients. BMC Cancer. 2010;10:511.
Sissung TM, Baum CE, Deeken J, et al. ABCB1 genetic variation influences the toxicity and clinical outcome of patients with androgen-independent prostate cancer treated with docetaxel. Clin Cancer Res. 2008;14(14):4543-9.
Sissung TM, Deeken J, Leibrand CR, et al. Identification of novel SNPs associated with risk and prognosis in patients with castration-resistant prostate cancer. Pharmacogenomics. 2016;17(18):1979-86.
Hahn NM, Marsh S, Fisher W, et al. Hoosier Oncology Group randomized phase II study of docetaxel, vinorelbine, and estramustine in combination in hormone-refractory prostate cancer with pharmacogenetic survival analysis. Clin Cancer Res. 2006;12(20 Pt 1):6094-9.
Derosa L, Galli L, Orlandi P, et al. Docetaxel plus oral metronomic cyclophosphamide: a phase II study with pharmacodynamic and pharmacogenetic analyses in castration-resistant prostate cancer patients. Cancer. 2014;120(24):3923-31.
de Morrée ES, Böttcher R, van Soest RJ, et al. Loss of SLCO1B3 drives taxane resistance in prostate cancer. Br J Cancer. 2016;115(6):674-81.
Takata R, Takahashi A, Fujita M, et al. 12 new susceptibility loci for prostate cancer identified by genome-wide association study in Japanese population. Nat Commun. 2019;10(1):4422.
Fantus RJ, Helfand BT. Germline genetics of prostate cancer: time to incorporate genetics into early detection tools. Clin Chem. 2019;65(1):74-9.