Corresponding author: Kinuko Ohneda, kohneda@megabank.tohoku.ac.jp, and Masayuki Yamamoto, masiyamamoto@med.tohoku.ac.jp
DOI: 10.31662/jmaj.2021-0156
Received: August 18, 2021
Accepted: December 1, 2021
Advance Publication: March 11, 2022
Published: April 15, 2022
Cite this article as:
Ohneda K, Hiratsuka M, Kawame H, Nagami F, Suzuki Y, Suzuki K, Uruno A, Sakurai-Yageta M, Hamanaka Y, Taira M, Ogishima S, Kuriyama S, Hozawa A, Tomita H, Minegishi N, Sugawara J, Danjoh I, Nakamura T, Kobayashi T, Yamaguchi-Kabata Y, Tadaka S, Obara T, Hishimuma E, Mano N, Matsuura M, Sato Y, Nakasone M, Honkura Y, Suzuki J, Katori Y, Kakuta Y, Masamune A, Aoki Y, Nakayama M, Kure S, Kinoshita K, Fuse N, Yamamoto M. A Pilot Study for Return of Individual Pharmacogenomic Results to Population-Based Cohort Study Participants. JMA J. 2022;5(2):177-189.
Introduction: Pharmacogenomic (PGx) testing results provide valuable information on drug selection and appropriate dosing, maximization of efficacy, and minimization of adverse effects. Although the number of large-scale, next-generation-sequencing-based PGx studies has recently increased, little is known about the risks and benefits of returning PGx results to ostensibly healthy individuals in research settings.
Methods: Single-nucleotide variants of three actionable PGx genes, namely, MT-RNR1, CYP2C19, and NUDT15, were returned to 161 participants in a population-based Tohoku Medical Megabank project. Informed consent was obtained from the participants after a seminar on the outline of this study. The results were sent by mail alongside sealed information letter intended for clinicians. As an exception, genetic counseling was performed for the MT-RNR1 m.1555A > G variant carriers by a medical geneticist, and consultation with an otolaryngologist was encouraged. Questionnaire surveys (QSs) were conducted five times to evaluate the participants’ understanding of the topic, psychological impact, and attitude toward the study.
Results: Whereas the majority of participants were unfamiliar with the term PGx, and none had undergone PGx testing before the study, more than 80% of the participants felt that they could acquire basic PGx knowledge sufficient to understand their genomic results and were satisfied with their potential benefit and use in future prescriptions. On the other hand, some felt that the PGx concepts or terminology was difficult to fully understand and suggested that in-person return of the results was desirable.
Conclusions: These results collectively suggest possible benefits of returning preemptive PGx information to ostensibly healthy cohort participants in a research setting.
Key words: Return of genomic results, Pharmacogenomics, Population-based cohort study
Individual genomic data collected in large-scale cohort studies are expected to be used as a tool for improving disease treatment and prevention and the management of personal healthcare. Pharmacogenomics is the study of individual genetic variability and how it influences drug response. The results of pharmacogenomic (PGx) testing provide valuable information on drug selection and appropriate dosing, maximization of efficacy, and minimization of adverse effects (1), (2). However, several considerations were raised when returning PGx results to individuals (3). For instance, the Industry Pharmacogenomics Working Group published “Points-to-Consider” related to the return of genomic results in the context of drug development (4), (5). Most concerns, including those related to ethical and legal considerations, clinical relevance, data quality control, and reporting results to participants and clinicians, are also applied in nonindustrial research and clinical settings. Although the number of large-scale, next-generation-sequencing-based PGx studies has increased [e.g., the Electronic Medical Records and Genomics Network (eMERGE-PGx) study (6)], little is known about the risks and benefits of returning PGx results to study participants. Moreover, potential differences between clinical and research settings might affect the outcome and impact on participants. In a clinical setting, patients are aware of the practical benefits of PGx testing as the results can be utilized by clinicians immediately after the genetic testing. On the contrary, the situation is different in returning preemptive PGx results in research settings (7), (8), (9). Population-based cohort study participants might not perceive any practical benefits from their PGx results. In addition, regardless of the elapsed time, patients should be informed of the PGx testing results by physicians so as to specify the correct treatment, thereby enabling patients to remember their relevance and the potential use of this data. Therefore, the potential benefits and challenges of returning PGx results should be evaluated.
The Tohoku Medical Megabank (TMM) project is part of a national project established in 2012 at Tohoku University and Iwate Medical University. We have conducted two large-scale, prospective cohort studies (the TMM Project Community-based Cohort Study and the TMM Project Birth and Three-generation Cohort Study) and developed an integrated biobank comprising biospecimens, multiomics data including whole-genome sequencing (WGS) data, and health information (10), (11), (12), (13), (14), (15), (16), (17). By using the WGS data, we accurately designed a system to return individual genomic results (return of genomic results: ROGR) to benefit the study participants for their personal healthcare management. Since the ROGR studies on ostensibly healthy individuals had been rarely conducted, we planned a series of pilot studies in a stepwise manner. The first pilot study was conducted to return genetic results associated with familial hypercholesterolemia to individuals with dyslipidemia. Since the participants had received their blood lipid data, receiving individual genomic information could be acceptable. A total of 215 individuals consented to participate in the study, of whom 23 were pathogenic variant carriers. This study was performed as the second pilot study. Preemptive PGx genotyping was performed for the study participants. In the third study, it was planned to return genomic information related to single-gene disorders with intermediate penetrance and/or adult-onset phenotype. The details of the TMM project ROGR pilot study are described elsewhere (18).
In the present study, we report our experience in returning individual PGx results to population-based genome cohort participants enrolled in the TMM project. In addition, we demonstrate that most of the participants felt that they could acquire basic PGx knowledge sufficient to understand their genomic results and were satisfied with their potential benefit and use in future prescriptions. These results suggest possible benefits of returning PGx results in a research setting.
All studies were conducted in accordance with the “Ethical guidelines for human genome and gene analysis research” presented by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Ministry of Health, Labor and Welfare (MHLW), and Ministry of Economy, Trade and Industry, as well as with the “Ethical guidelines for medical and health research involving human subjects” presented by MEXT and MHLW. The PGx pilot study was approved by the ToMMo Research Ethics Review Board of Tohoku University (approval code: 2019-4-094), and the study plan was reviewed by the Return of Genomic Results Review Committee (RGRRC).
Prior to the recruitment of the participants, the study plan was announced to physicians and pharmacists working in the Miyagi Prefecture as stakeholders for the study. The Pharmaceutical Department of Tohoku University Hospital (TUH) supported clinicians prescribing based on PGx results and assisting ToMMo staff in responding to local clinician inquiries. Furthermore, we held oral briefings at academic conferences for community and hospital pharmacists to inform the scientific community about the study and request for their cooperation. For local clinicians, the study announcement and request for cooperation were made using a mailing list. In addition, we prepared a leaflet targeting local clinicians in the form of this study and made it available at various academic conferences.
The flow diagram of the study design is presented in Figure 1. The individual IDs for the participants were securely converted into internal IDs for anonymized data (18), and two bioinformaticians dealing only with anonymized data independently performed variant detection using the WGS data. In the TMM project, WGS of the cohort participants was performed, and genome reference panels for the Japanese population were constructed. The genome reference panel has been updated with an increased number of participants with the WGS data (1KJPN, 2KJPN, 3.5KJPN, 4.7KJPN, and 8.3KJPN; Allele Frequency Panels)(19), (20). The present study used 4.7KJPN constructed from 4,773 Japanese individuals, of whom 4,378 were TMM cohort study participants.
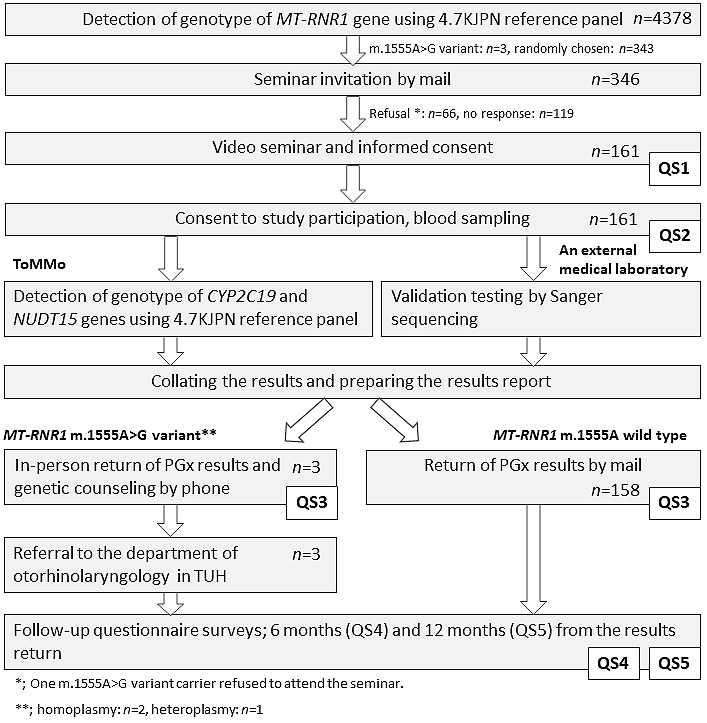
To detect individual genotypes, the pathogenic variant of the mitochondrial 12S ribosomal RNA, encoded by MT-RNR1 m.1555A>G [pathogenic variant (rs267606617), hereafter referred to as “m.1555A>G”], was searched against the file of the merged individual genotype of 4.7KJPN. Three participants were identified as carriers of the m.1555A>G variant and were recruited for the study. We evaluated the presence of the CYP2C19 variant *1-*3 (*1: wild type; *2: c.681G>A (p.Pro227=), rs4244285; and *3: c.636G>A (p.Trp212Ter), rs4986893) and NUDT15 codon 139 [c.415C>T (p.Arg139Cys), rs116855232; c.416G>A (p.Arg139His), rs147390019] only in the cohort participants that consented to participate in the study (n = 161). The participant genotypes were securely transferred to medical genetics experts through ID conversion. To validate the genomic results, the blood samples of the participants were sent to LSI Medience Corporation (LSI), a registered clinical laboratory that meets the standard policy by the Regulation for Enforcement of the Act on Clinical Laboratory Technicians governed by the MHLW. The results from LSI were collated with the ToMMo data. The m.1555G allele of a m.1555A>G heteroplasmic variant could not be detected using a variant panel of fixed genotypes for mitochondrial DNA, as the proportion of variant copies was <20% in this individual. The variant was detected by Sanger sequencing via LSI, which was confirmed by inspecting the proportion of sequence reads with the variant in the WGS data in ToMMo and further checked via LSI using the restriction fragment length polymorphism methods.
To evaluate the practical benefits to and psychological impact on the participants following the return, we conducted a series of QSs. In Table 1, the survey items are listed. We used three kinds of psychological testing: the Japanese version of the Kessler Psychological Distress Scale (K6) (21), the Japanese translation of the Profile of Mood States second edition (22), and the Japanese-language version of the Impact of Event Scale-Revised (23). The results of time-dependent changes in the psychological effects on the participants will be described separately. Eventually, four of nine subjects with high K6 scores underwent psychological assessment, although none left the study prematurely due to mental health problems.
Table 1. Questionnaire Items.
| Timing of survey | QS1 | QS2 | QS3 | QS4 | QS5 |
|---|---|---|---|---|---|
| Seminar invitation | Informed consent | Return of genomic results | 6 months from QS3 | 12 months from QS3 | |
| K6 | ● | ● | ● | ● | ● |
| POMS-2 | ● | ● | ● | ● | |
| IES-R | ● | ● | ● | ||
| Knowledge about PGx | ● | ● | ● | ||
| Intent of participation | ● | ● | |||
| Satisfaction with participation | ● | ● | ● |
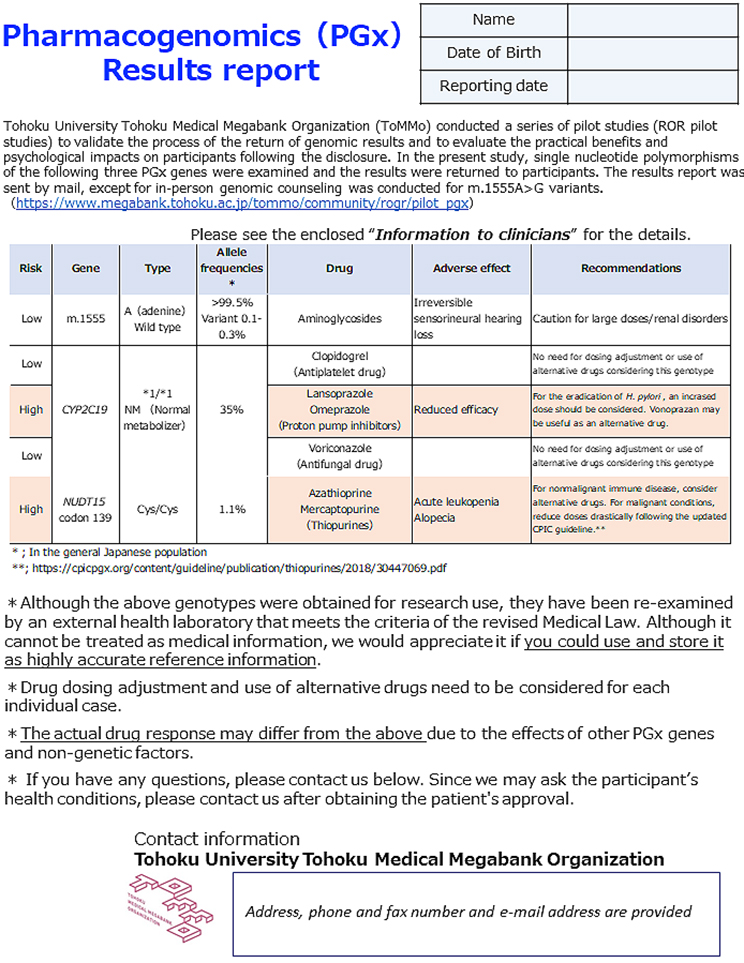
The PGx genes for return were selected according to the following criteria: (i) there was clear evidence of the genotype-phenotype associations for Japanese individuals; (ii) the results were clinically actionable; (iii) the frequency of the mutant alleles was not rare in the Japanese population; and (iv) the target drug was commonly prescribed in Japan. First, we examined the levels of evidence, guidelines, and actionability using PharmGKB (https://www.pharmgkb.org/), the Table of Pharmacogenomic Biomarkers in Drug Labeling by the US Food and Drug Administration (https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling), and Clinical Pharmacogenetics Implementation Consortium guidelines (https://cpicpgx.org/). Subsequently, we examined data from Japanese patients and selected three genes, namely, MT-RNR1, CYP2C19, and NUDT15 (Table 2). Drugs whose efficacy and safety are affected by the genotype (hereafter referred to as target drugs) are listed in Table 2. A previous study reported that the m.1555A>G pathogenic variant of MT-RNR1 in 1.9% of Japanese hearing-impaired patients (n = 264) diagnosed using the invader-based genetic screen test (24), whereas another report described that 1 in 1,683 (0.06%) individuals from the Japanese general population participating in the IWAKI Health Promotion Project was found to harbor the m.1555A>G variant according to TaqMan genotyping (25). Therefore, we searched this variant against the file of the merged individual 4.7KJPN genotype, which resulted in the recruitment of three variant carriers for the study. We selected the CYP2C19 polymorphism based on its serving as a predictive marker of the effectiveness of proton pump inhibitors (PPIs), lansoprazole, and omeprazole and because PPI-mediated eradication of Helicobacter pylori is a standard and preventive treatment for gastric cancer in Eastern Asia (26), (27). The Cys/Cys genotype of NUDT15 codon 139, which predicts the risk of thiopurine-induced severe adverse effects, is observed in ~1% of the Japanese population (28), (29), (30).
Table 2. Genes and Drugs Eligible for Disclosure.
| Gene | Drug | CPIC guidelines | CPIC level | PharmGKB level of evidence | FDA-approved drug label |
|---|---|---|---|---|---|
| MT-RNR1 m.1555 | Aminoglycosides | ||||
| CYP2C19 | Clopidogrel | Yes | A | 1A | Actionable |
| Voriconazole | Yes | A | 1A | Actionable | |
| Lansoprazole | Yes | A | 1A | Informative | |
| Omeprazole | Yes | A | 1A | Actionable | |
| NUDT15 | Azathioprine | Yes | A | 1A | Testing recommended |
| Mercaptopurine | Yes | A | 1A | Testing recommended | |
| Level Definitions for the CPIC Genes/Drugs are described in https://cpicpgx.org/prioritization/#flowchart. Clinical annotation levels of evidence for Pharm GKB are described in https://www.pharmgkb.org/page/clinAnnLevels. | |||||
A total of 346 primary participants recruited for the study were randomly selected from 4,378 cohort participants with the WGS data. All primary participants (i.e., three carriers of the m.1555A>G variant and 343 randomly chosen TMM cohort participants) were >20 years of age, with an average age of 61.3 and 64.3 years for study and nonstudy participants, respectively. The age and sex distributions of the participants are presented in Table 3. A recruitment letter was sent with information regarding the ROGR pilot study and a request for them to attend a video seminar. A total of 161 primary participants agreed to participate. A video seminar about the study was held for ~10 participants at a time in seven local assessment centers located in the Miyagi Prefecture (14). At the time of enrollment, none of the participants have taken the target drugs described in this study. Of the 161 seminar participants, 70 were male and 91 were female. Among the three subjects carrying the m.1555A>G variant, one did not consent to attend the seminar. The participants were asked to bring their prescription record notebook, a commonly used record of personal prescription history in Japan. At the seminar reception, the community support center staff interviewed the participants about their medical history.
Table 3. Age and Sex Distribution.
| Age group | Male | Female |
|---|---|---|
| 20–29 | 0 | 0 |
| 30–39 | 7 | 8 |
| 40–49 | 8 | 8 |
| 50–59 | 12 | 19 |
| 60–69 | 14 | 23 |
| 70–79 | 23 | 30 |
| 80–89 | 6 | 2 |
| 90–99 | 0 | 1 |
| Total | 70 | 91 |
The seminar participants were shown a video describing basic PGx concepts and the outline of this study for 10 min. Subsequently, a clinician provided supplementary information and answered questions raised by the seminar participants. Written informed consent was obtained from the seminar participants by the genome medical research coordinators, who underwent specialized training through ToMMo (31). All the seminar participants consented to participate in the study. Whole blood samples were taken from the participants and sent to an external registered clinical laboratory to conduct PGx gene sequencing so as to confirm the WGS results.
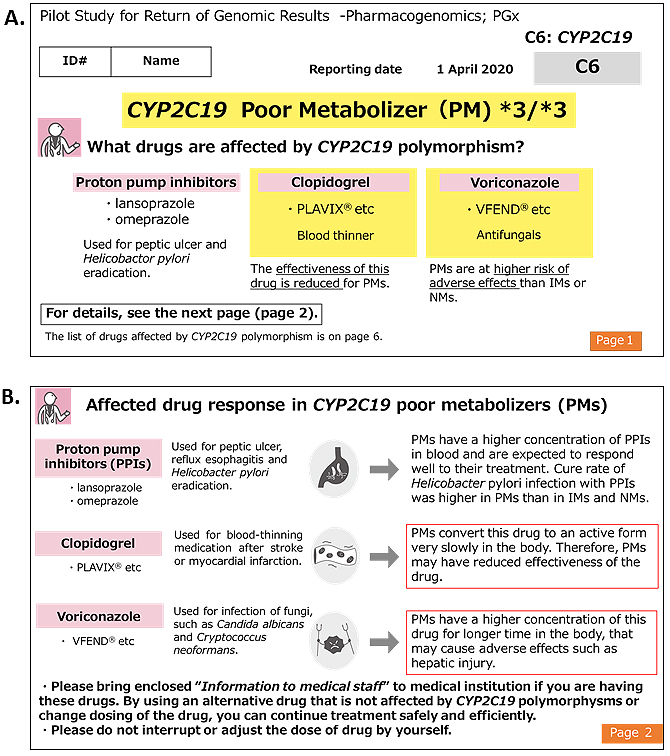
The genomic testing results provided by a registered clinical laboratory were collated with individual WGS data stored in the ToMMo. All the results matched, except for one case of a heteroplasmic m.1555A>G variant. The genotype results of the participants are presented in Table 4. The allele frequencies of CYP2C19 (32), (33) and NUDT15 (29) were consistent with those previously reported in Japanese individuals (ToMMo 4.7KJPN and a previous report (20)). The results report was sent to the participants by mail. Two types of reports were enclosed: one intended for the study participants and the other for healthcare professionals. The latter was prepared using two sets of sealed documents. Representative reports for the participants and healthcare professionals are presented in Figure 2 and 3, respectively. In the reports for the participants, the generic and brand names of the drugs considered as potential risks alongside their indication, medicinal effects, and individual predicted genotype-based response to the drug were informed. We asked the participants to bring the enclosed “information for healthcare professionals” to a hospital or pharmacy for any necessary treatment revision and to not interrupt or change the dose of the drug at their own discretion. In addition, we noted that drug-response prediction might not always be accurate due to individual variations in other PGx genes, and several nongenetic factors, such as food intake, liver and kidney functions, and concomitant drugs, could potentially affect drug response. Individual reports for healthcare professionals were prepared as a single sheet containing all three genes, which makes them manageable as supporting medical information. These reports include medication and dosage recommendations based on the genomic results. In addition to the results report, information on the ROGR pilot studies, PGx results interpretation, and a URL linked to PGx guidelines and databases were enclosed. The participants could request additional sets of “information for healthcare professionals” as needed and contact the researcher for additional information.
As described in the Methods section, one participant harbored a heteroplasmic m.1555A>G variant with m.1555A as the dominant type. Because the heteroplasmic variant frequency varies according to tissue type (34), we returned the genomic results individually according to the same flow employed for carriers of the homoplasmic m.1555A>G variant.
Due to COVID-19 pandemic constraints, genetic counseling and return of the PGx test results for the carriers of the m.1555A>G variant were performed by phone, with the results report sent via mail in advance. In addition to the information on the drug-induced hearing loss, the results report included an example family tree demonstrating maternal inheritance and a list of aminoglycosides used in Japan. In addition, two sets of a medication alert card for aminoglycosides in Japanese and English were provided. Prior to genetic counseling, we informed the participants that a family history of hearing loss would be discussed. Genetic counseling was performed by a medical geneticist. The profile of the m.1555A>G variant carriers is presented in Table 5. They were all females in the 70s. Case 1 has a hearing loss in her left ear that occurred in her 40s, although her medication history of aminoglycosides is uncertain. Case 2 does not have a hearing impairment. Both cases have siblings diagnosed with drug-induced hearing loss without genetic testing. Case 3 is a carrier of the heteroplasmy variant who does not have past medical nor family history of hearing impairment. After ~30 min of genetic counseling performed by a clinical geneticist, all the participants carrying the m.1555A>G variant (n = 3; two homoplasmic and one heteroplasmic) consented to visit an otolaryngologist in TUH, for which we prepared a medical information provision form with the results of the PGx testing and a genetic family tree. Since all of them shared the results with relatives who are at risk, we informed them that they could visit TUH and have a genetic counseling with an otolaryngologist. After the examination, the attending otolaryngologist entered aminoglycosides as contraindicated drugs in the participant electronic healthcare records (EHRs) in TUH.
Table 5. Profiles of the MT-RNR1 m.1555A > G Variant Carriers.
| Case # | Age | Sex | Genotype | Presence of hearing loss | |
|---|---|---|---|---|---|
| Study participant | Families | ||||
| 1 | 70s | Female | Homoplasmy | Yes | Yes |
| 2 | 70s | Female | Homoplasmy | No | Yes |
| 3 | 70s | Female | Heteroplasmy | No | No |
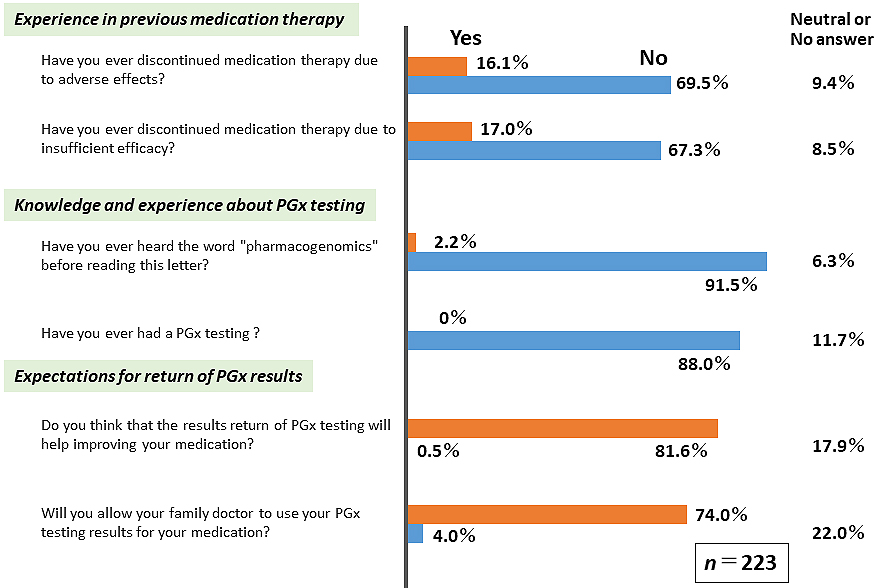
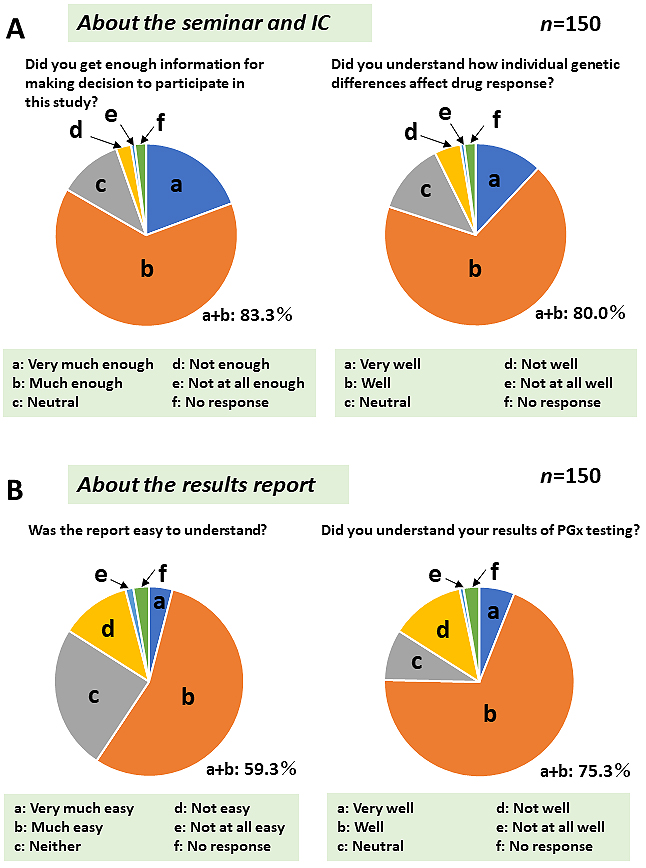
Because detailed information regarding the PGx field is not generally accessible to a broader audience, we provided participants with basic PGx information several times throughout the study period. Questionnaire surveys (QSs) were conducted five times before (QS1 and QS2, Figure 4) and after (QS3, QS4, and QS5, Figure 5) the ROGR. The results indicated that only a small percentage of participants (<17%) had previous problems with medication therapy. As expected, majority of the participants were unfamiliar with the term PGx, and none had undergone PGx testing before the study. However, after reading the recruitment letter, >70% of the participants expected that the PGx testing results might be beneficial (Figure 4). Upon receiving the PGx results, majority (>80%) of the participants felt that they could obtain sufficient information in the seminar and the IC form (Figure 5A). However, only a small percentage of the participants felt that the results report was easy to understand, whereas the majority reported having only a moderate understanding of the PGx testing results (Figure 5B). In the free description in QS4, some participants suggested that in-person return of the results was desirable. These results indicate a limitation in the participants’ comprehension of the returned genomic results without an in-person interaction.
We conducted a pilot study with regard to the return of three PGx results to participants enrolled in a large-scale genome cohort TMM project. We identified three carriers of the m.1555A>G variant, and they were subsequently referred to the otorhinolaryngology department in TUH for further testing and counseling. Genetic counseling was successfully performed by a clinical geneticist by phone in addition to return of the results report prior to genetic counseling. Most of the participants reported that their knowledge and understanding of the PGx testing have improved after receiving the results report. Given that large-scale genome cohort studies have been increasingly conducted for healthy individuals in research settings, this study provides key insights into the potential use of PGx information in personalized healthcare management.
However, some challenges and limitations remain. First, we could only return a limited number of PGx genes. We considered the inclusion of the CYP2D6 polymorphism, as this gene predicts the risk of severe adverse effects when using codeine (35); however, it was excluded, because the number of carriers with a higher risk of poor treatment outcomes was much lower in individuals with Japanese rather than European ancestry (36). In addition, genotype confirmation is difficult for such a large number of subjects and would merit a separate study to better focus on participant education with regard to this specific gene. To scale up the number of PGx genes and participants, the cost-effectiveness of preemptive PGx testing should be evaluated and the scalability of human resources enhanced. Second, we encountered challenges in the process of confirming the m.1555A>G heteroplasmic variant, as described in the Methods section. We could detect the genotype by collating the genomic results independently obtained by two different laboratories. This suggests the importance of the validation testing because mismatch of the genotyping results could also occur due to human error during handling of the samples. Third, as reported in the Return of Actionable Variant Empirical (RAVE) study conducted at the Mayo Clinic (37), we encountered participant contact-related challenges. Approximately 33% (n = 119) of the 346 subjects invited to participate in the video seminar did not respond to the invitation letter; thus, a second invitation was sent, followed by attempts to contact the subjects by phone. We needed to call more than three times to contact some subjects who responded that they would attend the seminar. These experiences indicate that a considerable amount of time and human resources are required for successful participant recruitment. Fourth, even though the basic information of PGx had been given several times, some participants felt that the PGx concepts or terminology was difficult to fully understand. To accurately evaluate and take measures for improvement of the participants’ understanding, objective testing is necessary.
Although this study showed possible benefits of the return of PGx results individually in a research setting, further challenges and limitations hinder clinical implementation in Japan. In Japanese medical institutions, PGx testing is conducted in patients with an existing disease diagnosis. However, the Japanese health insurance currently covers PGx testing for only two genes, namely, the UGT1A1 polymorphism, which predicts the risk of severe adverse effects associated with the administration of irinotecan (38), and the NUDT15 polymorphism, which was examined in this study. We added the health insurance information in the results report for a NUDT15 Cys/Cys variant, because the insurance coverage began in February 2019 during this study. With regard to the m.1555A>G variant data, medical implementation was performed by entering the contraindicated drug information in the appropriate EHRs of TUH. Nevertheless, the Japanese EHR system is currently under development to include the use of the PGx data. To clinically implement PGx data considerations for a large number of undiagnosed patients, a multicenter study, such as the one employed to design the eMERGE-PGx study (6), (39), (40), needs to be conducted in the near future. Because the TMM project was conducted involving population-based cohort participants, the PGx results must be submitted separately to healthcare professionals. Therefore, we prepared two sets of sealed documents with contact information. To improve information accessibility, it would be better to include the results of the Miyagi Medical and Welfare Information Network (MMWIN), a medical network system that provides data storage services for medical and pharmaceutical institutes (41). This system was established in 2013 to protect medical data from destruction by natural disasters, such as the Great East Japan Earthquake. Because MMWIN enables sharing of patient information, including contraindications, entry of such information by healthcare professionals could potentially help in preventing adverse drug effects. An update and further expansion of the information provided to users are worthy of further consideration to promote personalized medicine.
In summary, we described the possible benefits and challenges of returning PGx results via a large-scale integrated research biobank. The limitations experienced in conducting this research might help in the planning of other ROGR studies. The practical effectiveness of returning PGx data needs to be further evaluated.
None
This work was supported by the MEXT TMM project and the Japan Agency of Medical Research Development [AMED, JP 20 km0105001, and JP 20 km0105002].
We thank the chairperson Yoshimitsu Fukushima and members of the RGRRC for their critical comments and suggestions, Drs. Akimune Fukushima and Tomoharu Tokutomi for their collaboration with the ROGR pilot study, and Drs. Shin-ichi Usami and Shin-ya Nishio for their valuable suggestions regarding the m.1555A>G variant. We also thank Dr. Evelyn Marie Gutiérrez Rico and Editage (www.editage.com) for English language editing, Miho Kuriki for figure illustration, and Michiko Sugawara and Yoko Haga for secretarial assistance. The list of participating ToMMo members is available at https://megabank.tohoku.ac.jp/english/a201201.
Conceptualization: K.O., M.H., H.K., F.N., N.F., M.Y. Data Curation: S.O., I.D., T.N., M.N. Formal analysis: Yo.S., S.T., T.O., E.H., Y.Y-K. Funding acquisition: M.Y. Investigation: K.S., A.U., M.S-Y., M.M., Yu.S., M.N., M.T., H.T., Y.Ho., J.S., Yo K., Methodology: M.H., F.N. Project administration: K.O., M.H., H.K., N.F. Resources: S.K., A.H., N.M., J.S., T.K. Supervision: M.S., N.M., Yu K., A.M., Y.A., S.K., K.K., M.Y. Validation: Y.Y-K., S.T. Visualization: K.O., M.H., F.N., Y.Ha. Writing―original draft: K.O. Writing―review & editing: K.O., M.H., H.K., F.N., Yo.S., N.F., Y.Y-K., Y.Ho., Yo K., M.N., M.Y.
The project protocols were reviewed and approved by the Ethics Committee of Tohoku University Tohoku Medical Megabank Organization (IRB approval code number: 2019-4-094).
Weinshilboum RM, Wang L. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. 2017;92(11):1711-22.
Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394(10197):521-32.
Dressler LG. Return of research results from pharmacogenomic versus disease susceptibility studies: what’s drugs got to do with it? Pharmacogenomics. 2012;13(8):935-49.
Renegar G, Webster CJ, Stuerzebecher S, et al. Returning genetic research results to individuals: points-to-consider. Bioethics. 2006;20(1):24-36.
Prucka SK, Arnold LJ, Brandt JE, et al. An update to returning genetic research results to individuals: perspectives of the industry pharmacogenomics working group. Bioethics. 2015;29(2):82-90.
Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96(4):482-9.
Olson JE, Rohrer Vitek CR, Bell EJ, et al. Participant-perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time). Genet Med. 2017;19(7):819-25.
Lemke AA, Hulick PJ, Wake DT, et al. Patient perspectives following pharmacogenomics results disclosure in an integrated health system. Pharmacogenomics. 2018;19(4):321-31.
Meagher KM, Curtis SH, Borucki S, et al. Communicating unexpected pharmacogenomic results to biobank contributors: a focus group study. Patient Educ Couns. 2021;104(2):242-9.
Fuse N, Sakurai-Yageta M, Katsuoka F, et al. Establishment of integrated biobank for precision medicine and personalized healthcare: the Tohoku Medical Megabank project. JMA J. 2019;2(2):113-22.
Kuriyama S, Yaegashi N, Nagami F, et al. The Tohoku Medical Megabank project: design and mission. J Epidemiol. 2016;26(9):493-511.
Kuriyama S, Metoki H, Kikuya M, et al. Cohort profile: Tohoku Medical Megabank project birth and three-generation cohort study (TMM BirThree Cohort Study): rationale, progress and perspective. Int J Epidemiol. 2020;49(1):18-19m.
Minegishi N, Nishijima I, Nobukuni T, et al. Biobank establishment and sample management in the Tohoku Medical Megabank project. Tohoku J Exp Med. 2019;248(1):45-55.
Hozawa A, Tanno K, Nakaya N, et al. Study profile of the Tohoku Medical Megabank community-based cohort study. J Epidemiol. 2021;31(1):65-76.
Yasuda J, Kinoshita K, Katsuoka F, et al. Genome analyses for the Tohoku Medical Megabank project towards establishment of personalized healthcare. J Biochem. 2019;165(2):139-58.
Koshiba S, Motoike I, Saigusa D, et al. Omics research project on prospective cohort studies from the Tohoku Medical Megabank project. Genes Cells. 2018;23(6):406-17.
Sugawara J, Ishikuro M, Obara T, et al. Maternal baseline characteristics and perinatal outcomes: the Tohoku Medical Megabank project birth and three-generation cohort study. J Epidemiol. Forthcoming. 2021.
Kawame H, Fukushima A, Fuse N, et al. The return of individual genomic results to research participants: design and pilot study of Tohoku Medical Megabank Project. J Hum Genet. 2022;67:9-17.
Nagasaki M, Yasuda J, Katsuoka F, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018.
Tadaka S, Katsuoka F, Ueki M, et al. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6:28.
Kessler RC, Andrews G, Colpe LJ, et al. Short screening scales to monitor population prevalences and trends in non-specific psychological distress. Psychol Med. 2002;32(6):959-76.
Yokoyama KC. Production of Japanese edition of Profile of Mood States (POMS): assessment of reliability and validity. Jpn J Public Health. 1990;37(11):913-8.
Asukai N, Kato H, Kawamura N, et al. Reliability and validity of the Japanese-language version of the impact of event scale-revised (IES-R-J): four studies of different traumatic events. J Nerv Ment Dis. 2002;190(3):175-82.
Usami S, Nishio SY, Nagano M, et al. Simultaneous screening of multiple mutations by invader assay improves molecular diagnosis of hereditary hearing loss: a multicenter study. PLoS One. 2012;7(2):e31276.
Maeda Y, Sasaki A, Kasai S, et al. Prevalence of the mitochondrial 1555 A>G and 1494 C>T mutations in a community-dwelling population in Japan. Hum Genome Var. 2020;7:27.
Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20(16):4483-90.
Fallone CA, Chiba N, van Zanten SV, et al. The Toronto consensus for the treatment of helicobacter pylori infection in adults. Gastroenterology. 2016;151(1):51-69.e14.
Kakuta Y, Naito T, Onodera M, et al. NUDT15 R139C causes thiopurine-induced early severe hair loss and leukopenia in Japanese patients with IBD. Pharmacogenomics J. 2016;16(3):280-5.
Kakuta Y, Kinouchi Y, Shimosegawa T. Pharmacogenetics of thiopurines for inflammatory bowel disease in East Asia: prospects for clinical application of NUDT15 genotyping. J Gastroenterol. 2018;53(2):172-80.
Kakuta Y, Kawai Y, Okamoto D, et al. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: a multicenter study. J Gastroenterol. 2018;53(9):1065-78.
Sakurai-Yageta M, Kawame H, Kuriyama S, et al. A training and education program for genome medical research coordinators in the genome cohort study of the Tohoku Medical Megabank Organization. BMC Med Educ. 2019;19(1):297.
Tsuneoka Y, Fukushima K, Matsuo Y, et al. Genotype analysis of the CYP2C19 gene in the Japanese population. Life Sci. 1996;59(20):1711-5.
Ieiri I, Higuchi S. Pharmacogenetics of CYP2C subfamily in a Japanese population. J Toxicol Sci. 1998;23(Suppl 2):129-31.
del Castillo FJ, Rodríguez-Ballesteros M, Martín Y, et al. Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40(8):632-6.
Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95(4):376-82.
Dorji PW, Tshering G, Na-Bangchang K. CYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in South-East and East Asian populations: a systematic review. J Clin Pharm Ther. 2019;44(4):508-24.
Kochan DC, Winkler E, Lindor N, et al. Challenges in returning results in a genomic medicine implementation study: the Return of Actionable Variants Empirical (RAVE) study. NPJ Genom Med. 2020;5:19.
Innocenti F, Ratain MJ. “Irinogenetics” and UGT1A: from genotypes to haplotypes. Clin Pharmacol Ther. 2004;75(6):495-500.
Crosslin DR, Robertson PD, Carrell DS, et al. Prospective participant selection and ranking to maximize actionable pharmacogenetic variants and discovery in the eMERGE Network. Genome Med. 2015;7(1):67.
Rasmussen-Torvik LJ, Almoguera B, Doheny KF, et al. Concordance between research sequencing and clinical pharmacogenetic genotyping in the eMERGE-PGx Study. J Mol Diagn. 2017;19(4):561-6.
Ido K, Nakamura N, Nakayama M. Miyagi medical and welfare information network: A backup system for patient clinical information after the Great East Japan Earthquake and Tsunami. Tohoku J Exp Med. 2019;248(1):19-25.